-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00010825
MPSGTPAHHRHATPKPTVVERPHGHKEAPPQVIPMTNVMPGDSSFVTWDQFQFTMSVIRT
VIGVTVESGATIPILPGDSSTMQAFLLRIEPRYTQMGLEPREWGNALTDHLVGPALRYWM
YLRRTIDQSDWATVQRRLLERFDKTMSQSQLSTELAKVRWNGNPKEYTDRFVRWNGKPKE
YTDRFAAVTERGLGLAPDELADYYCTGLPTDLHLLITNNGQVKYQSWEQAATAATRLYEP
K
- Download Fasta
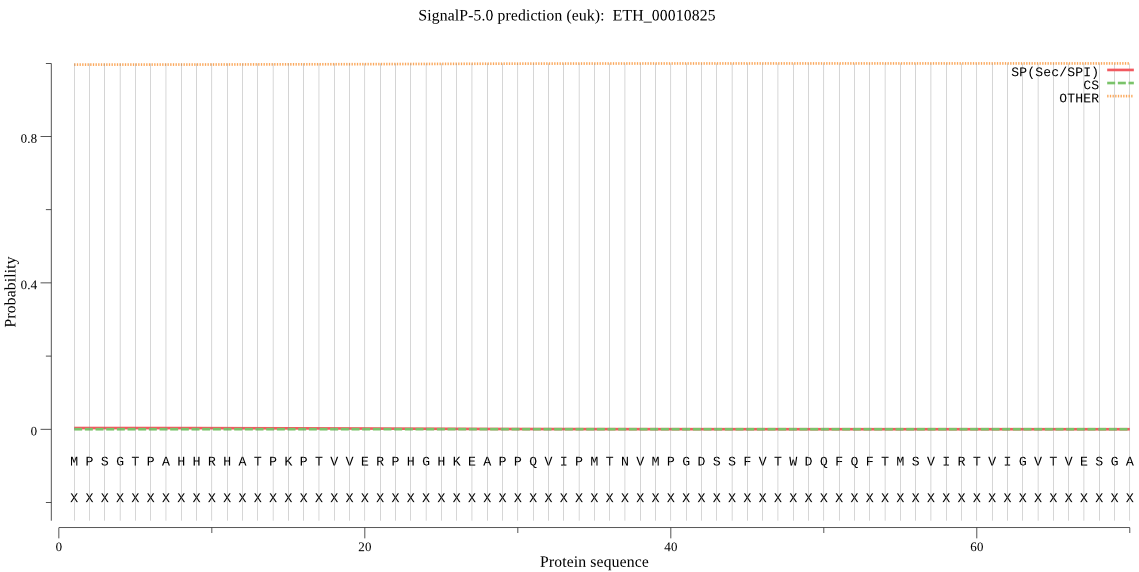
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00010825.fa
Sequence name : ETH_00010825
Sequence length : 241
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.018
CoefTot : -1.563
ChDiff : 1
ZoneTo : 19
KR : 2
DE : 0
CleavSite : 12
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.124 1.271 0.166 0.581
MesoH : -0.088 0.277 -0.291 0.235
MuHd_075 : 23.861 10.371 5.673 4.615
MuHd_095 : 12.220 6.801 1.816 2.208
MuHd_100 : 6.227 6.885 1.621 2.376
MuHd_105 : 3.167 6.388 2.276 2.168
Hmax_075 : 5.800 2.400 -1.219 1.660
Hmax_095 : 4.550 0.262 -2.570 1.470
Hmax_100 : 4.000 -0.500 -2.854 1.460
Hmax_105 : 3.733 2.000 -1.439 2.510
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9522 0.0478
DFMC : 0.9377 0.0623
-
Fasta :-
>ETH_00010825
ATGCCGAGCGGGACGCCAGCCCATCATCGCCATGCTACTCCGAAGCCCACCGTGGTAGAG
CGGCCGCATGGACATAAGGAAGCGCCCCCGCAAGTGATACCTATGACCAATGTTATGCCA
GGGGACAGTTCCTTCGTCACGTGGGACCAATTCCAGTTCACTATGAGTGTGATAAGAACT
GTTATAGGGGTGACCGTGGAATCCGGCGCAACGATTCCCATACTACCAGGAGACAGCAGC
ACTATGCAGGCCTTTCTGCTGCGCATCGAGCCCCGGTACACCCAGATGGGATTGGAACCG
CGCGAATGGGGAAATGCACTTACAGACCACCTTGTGGGCCCGGCTCTAAGGTATTGGATG
TATCTACGGAGAACTATCGACCAAAGCGACTGGGCGACAGTACAGCGCAGGCTTTTGGAG
CGGTTCGACAAAACAATGTCGCAGAGTCAATTGTCAACGGAGTTGGCTAAAGTACGGTGG
AATGGTAACCCGAAGGAATACACGGACCGGTTCGTACGGTGGAATGGTAAGCCGAAGGAA
TACACGGACCGGTTCGCGGCTGTAACGGAGCGCGGTTTGGGTCTTGCCCCCGACGAACTC
GCTGACTATTACTGCACCGGGCTACCGACAGACCTCCATCTTCTAATAACCAATAACGGA
CAGGTGAAATATCAGTCATGGGAACAAGCGGCGACAGCCGCAACACGGCTCTACGAGCCC
AAGTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00010825 | 13 T | HRHATPKPT | 0.996 | unsp | ETH_00010825 | 152 S | QSQLSTELA | 0.994 | unsp |


