

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
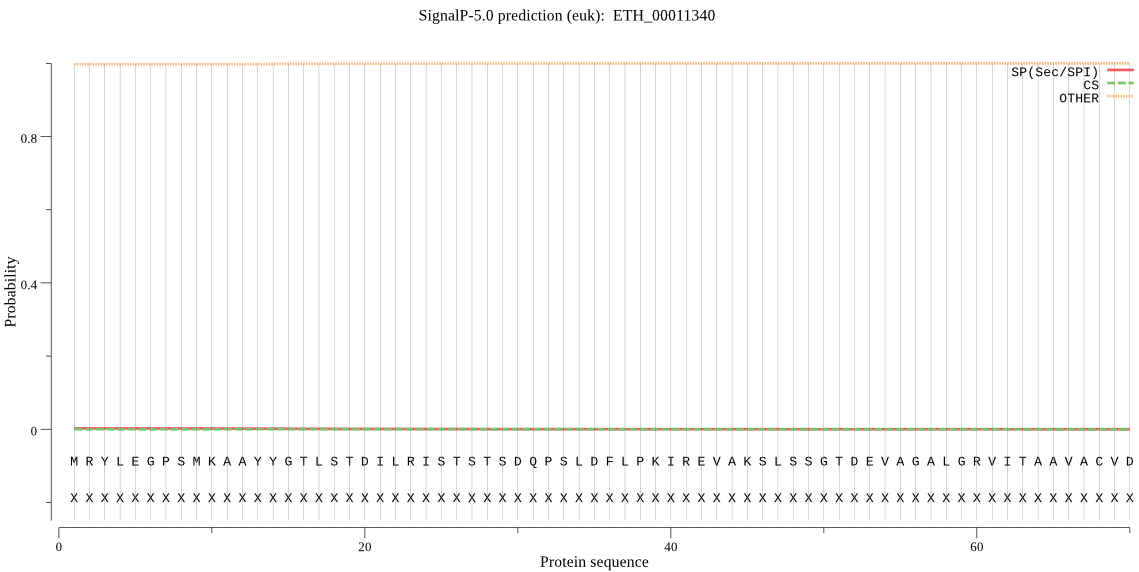
| ETH_00011340 | OTHER | 0.991307 | 0.002049 | 0.006644 |
MRYLEGPSMKAAYYGTLSTDILRISTSTSDQPSLDFLPKIREVAKSLSSGTDEVAGALGR VITAAVACVDKISLDRKVDVQRLGSLATNSQGNMSLQEPIKGTVNDGATSPQLLRAPSGL LGDPFQMRQWYLHSFFGNFTVEADRTWRKLEATENTKPVVLAILDTGCYLHQDFIDDFDI AKSIFWDNAGETDCSDGIDNDGNGYIDDCFGWNFVEDNGHPFTDDSGHGTSVTSVAAARA HDGKGGRGVLPNPTVMCLRVGSERGVWTSATIPALDYAVKMKARVSNHSYGGPG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00011340 | 27 S | RISTSTSDQ | 0.994 | unsp | ETH_00011340 | 49 S | KSLSSGTDE | 0.996 | unsp |