-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00011650
MPSGKPAHHRHATPKPTVAERPHGHKEGPPQVISMTNVMPGGSPFVTRDQFQSAMSVIRT
VIGGTVESGATIPILPEDGSTMQAFLLHIERRYTQMGLEPREWRNALIDHLVEDCVSDRL
KRSP
- Download Fasta
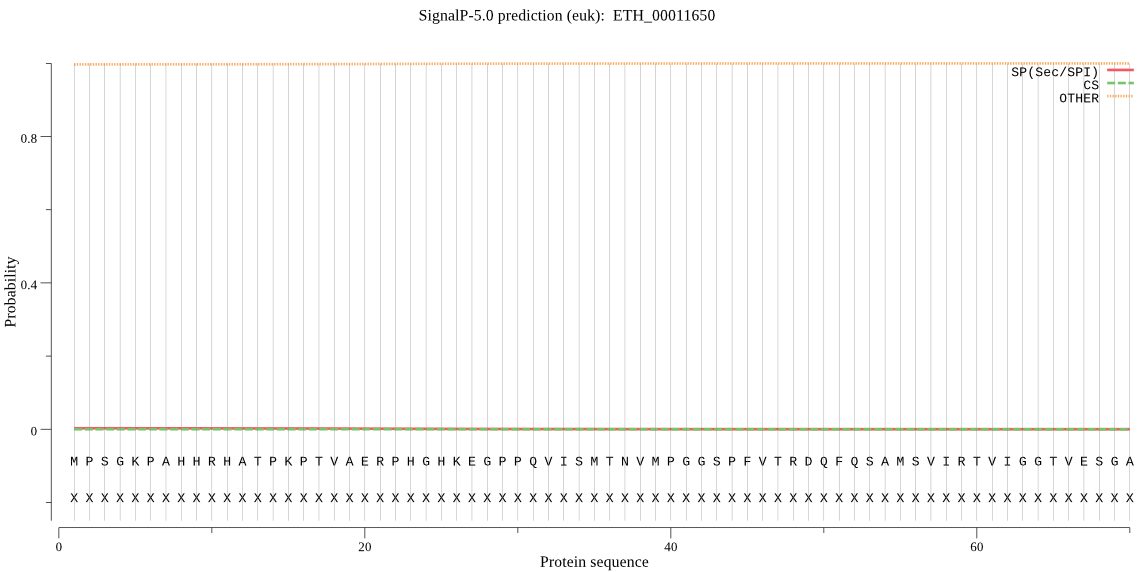
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00011650.fa
Sequence name : ETH_00011650
Sequence length : 124
VALUES OF COMPUTED PARAMETERS
Coef20 : 2.905
CoefTot : -1.535
ChDiff : 1
ZoneTo : 19
KR : 3
DE : 0
CleavSite : 12
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.041 0.982 0.118 0.514
MesoH : -0.311 0.121 -0.349 0.188
MuHd_075 : 33.688 13.811 8.430 6.061
MuHd_095 : 3.058 7.077 1.808 1.987
MuHd_100 : 5.299 7.176 3.846 1.643
MuHd_105 : 8.676 6.931 5.144 1.695
Hmax_075 : 5.800 2.400 -1.219 1.660
Hmax_095 : -4.025 -1.838 -2.901 0.753
Hmax_100 : -6.200 -2.900 -2.865 0.540
Hmax_105 : 3.733 2.000 -1.439 2.510
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9390 0.0610
DFMC : 0.9054 0.0946
-
Fasta :-
>ETH_00011650
ATGCCGAGCGGGAAGCCAGCCCATCATCGCCATGCTACTCCGAAGCCTACAGTCGCAGAG
CGGCCGCATGGGCATAAGGAGGGGCCCCCGCAGGTGATATCTATGACCAATGTTATGCCA
GGGGGCAGTCCCTTCGTCACGAGGGACCAATTCCAGTCCGCTATGAGTGTGATAAGAACT
GTTATAGGCGGGACCGTGGAATCGGGCGCAACAATTCCCATACTACCAGAAGACGGCAGC
ACTATGCAGGCCTTTCTGCTGCACATCGAGCGCCGGTACACCCAGATGGGATTGGAGCCG
CGCGAGTGGAGGAATGCACTTATTGACCACCTTGTGGAGGATTGTGTTTCAGACCGTCTG
AAGCGCAGCCCTTGA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00011650 | 13 T | HRHATPKPT | 0.996 | unsp | ETH_00011650 | 56 S | QSAMSVIRT | 0.996 | unsp |


