

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00012075 | OTHER | 0.999996 | 0.000004 | 0.000000 |
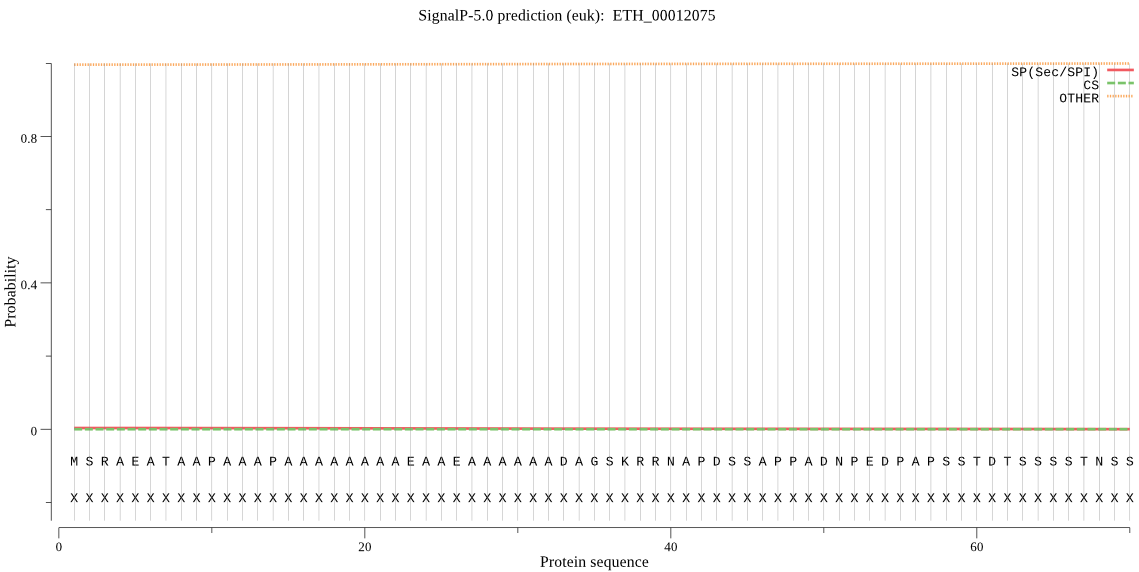
MSRAEATAAPAAAPAAAAAAAAEAAEAAAAAADAGSKRRNAPDSSAPPADNPEDPAPSST DTSSSSTNSSSSSSSSSSSSSSSSSSSTAKRVRRTCPYLGTINKHMLDFDFEKVCSICLS NQHVYACLVCGRYFQGRGKSTFAFMHALEQRHFVYVNLTTCKVYCLPDNYEVQDYSLQDI IYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPL RNYLLLLDLSEKQKYNSVLKTLSELMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGE QSDPLALFAWLLNVLHRDQKDKKTGGSIIHDCFQGEVLVTDAQSEEGRKTPFLYLTLDLP PAPIFKDSLDRNMIPTVPIFDLLQKFDGETDSGPTPLQRKKCSLYKLPRYLVLAVKRFSK NNFFVEKNPTIVTFPVKNLDLREYVHPDALEDNPVTRYNLVANICHQGKPHDGFYKAQVL HAPTNEWHEMEDLRVTPVLPQFVALSEAYIQFYQRQDVMADGSLDEALQQKCLQEMKKVK TQAEPEDEIEDIFAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00012075 | 74 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 74 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 74 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 75 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 76 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 77 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 78 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 79 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 80 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 81 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 82 S | SSSSSSSSS | 0.99 | unsp | ETH_00012075 | 84 S | SSSSSSSST | 0.993 | unsp | ETH_00012075 | 58 S | DPAPSSTDT | 0.996 | unsp | ETH_00012075 | 72 S | NSSSSSSSS | 0.994 | unsp |