-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
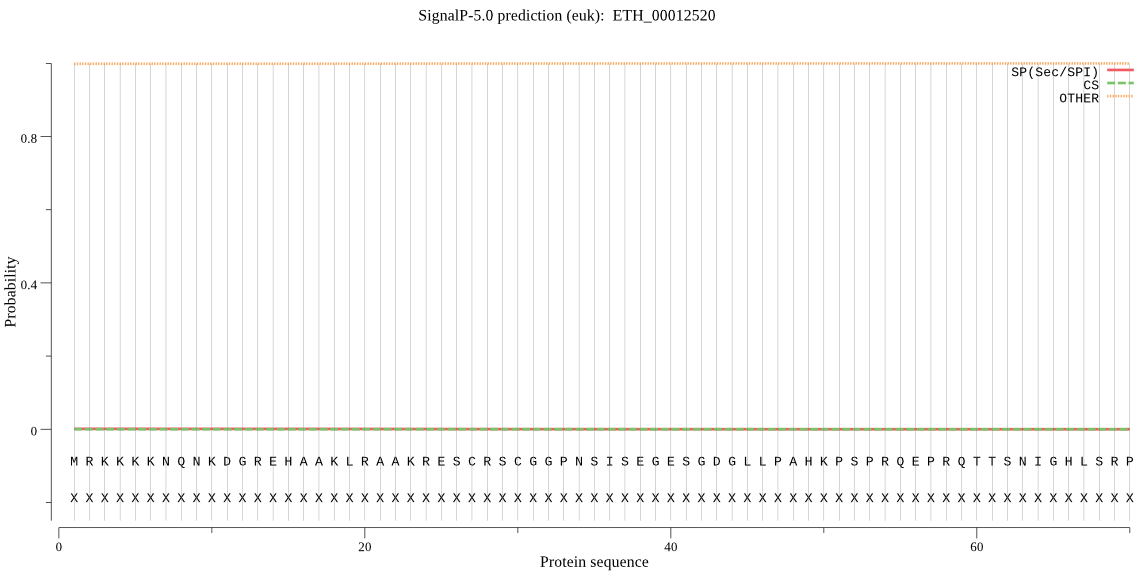
ETH_00012520 | OTHER | 0.999986 | 0.000004 | 0.000010 | |
No Results
-
Fasta :-
>ETH_00012520
MRKKKKNQNKDGREHAAKLRAAKRESCRSCGGPNSISEGESGDGLLPAHKPSPRQEPRQT
TSNIGHLSRPTITALVHGLNRNYYSIAINCRKGELEHQMLANLHTNKWSDALKLKNYQEH
QEEREESIAALKELSCRYTSMIEEEIKKKPEQLLVDRAGQIDAKMRIQQTLDQLMTDTLL
QSLGSMISTLVF
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00012520.fa
Sequence name : ETH_00012520
Sequence length : 192
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.514
CoefTot : -0.853
ChDiff : 7
ZoneTo : 10
KR : 6
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.006 0.982 0.011 0.399
MesoH : -1.481 -0.303 -0.607 -0.044
MuHd_075 : 17.213 10.097 5.559 3.796
MuHd_095 : 36.967 14.583 9.527 6.400
MuHd_100 : 26.617 9.495 6.671 4.514
MuHd_105 : 10.639 2.704 2.941 1.998
Hmax_075 : -24.063 -9.012 -8.055 -2.905
Hmax_095 : -18.700 -9.012 -7.627 -1.910
Hmax_100 : -14.700 -10.800 -8.424 -1.790
Hmax_105 : -28.600 -14.600 -11.077 -4.160
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9516 0.0484
DFMC : 0.8365 0.1635
-
Fasta :-
>ETH_00012520
ATGAGAAAGAAGAAAAAGAATCAAAATAAGGACGGACGTGAACACGCAGCAAAGCTTCGA
GCAGCTAAACGCGAGAGCTGTCGGAGTTGTGGTGGACCCAATTCAATCAGTGAAGGGGAA
AGTGGTGATGGACTGCTTCCGGCTCATAAACCCTCACCTCGACAGGAGCCGCGGCAGACA
ACTTCGAACATTGGCCACTTGTCGCGCCCCACAATAACAGCTCTGGTCCACGGACTTAAC
CGAAATTATTATTCAATTGCAATTAACTGCCGCAAAGGCGAACTCGAACACCAAATGCTG
GCCAACCTGCACACCAACAAGTGGAGCGACGCCCTCAAACTCAAGAACTACCAAGAGCAC
CAGGAGGAGCGGGAGGAAAGCATCGCGGCGCTGAAGGAGCTGAGCTGTCGATACACCTCA
ATGATTGAAGAAGAAATAAAAAAGAAACCCGAACAACTTCTCGTTGACAGAGCTGGACAA
ATAGATGCAAAAATGCGGATTCAGCAAACCCTAGATCAGCTGATGACGGACACGCTGCTG
CAGTCCCTCGGCAGCATGATCTCCACGCTCGTCTTCTAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00012520 | 52 S | AHKPSPRQE | 0.991 | unsp | ETH_00012520 | 140 S | CRYTSMIEE | 0.997 | unsp |


