

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00013695 | OTHER | 0.957589 | 0.002083 | 0.040328 |
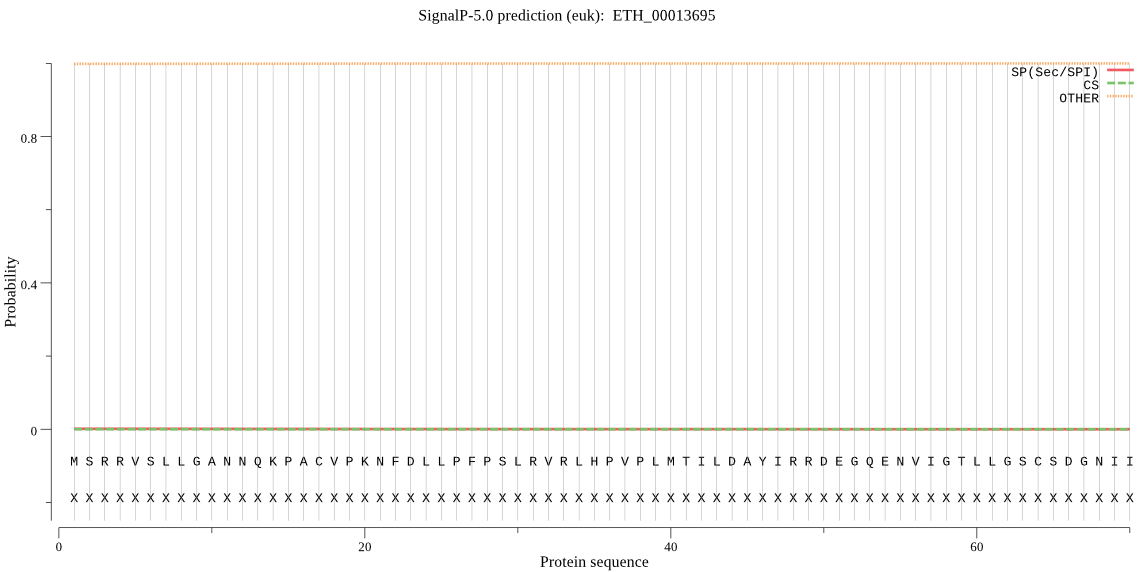
MSRRVSLLGANNQKPACVPKNFDLLPFPSLRVRLHPVPLMTILDAYIRRDEGQENVIGTL LGSCSDGNIIDVTDCFVDRHSLTGEGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWF CTGSEMTELTCAVHGWFKQFSSVSKFFPQPPLTEPIHLMVDAVMESESFSVKVYTQVQMT MAREACFQFHELPLELYATPSDRVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDIT DGLNAELEQLQEKLEICAAYVRRVLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQN TLQDSLMAAHLARLAKLQFAVAQKLNTSFF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| ETH_00013695 | 81 S | VDRHSLTGE | 0.991 | unsp |