

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
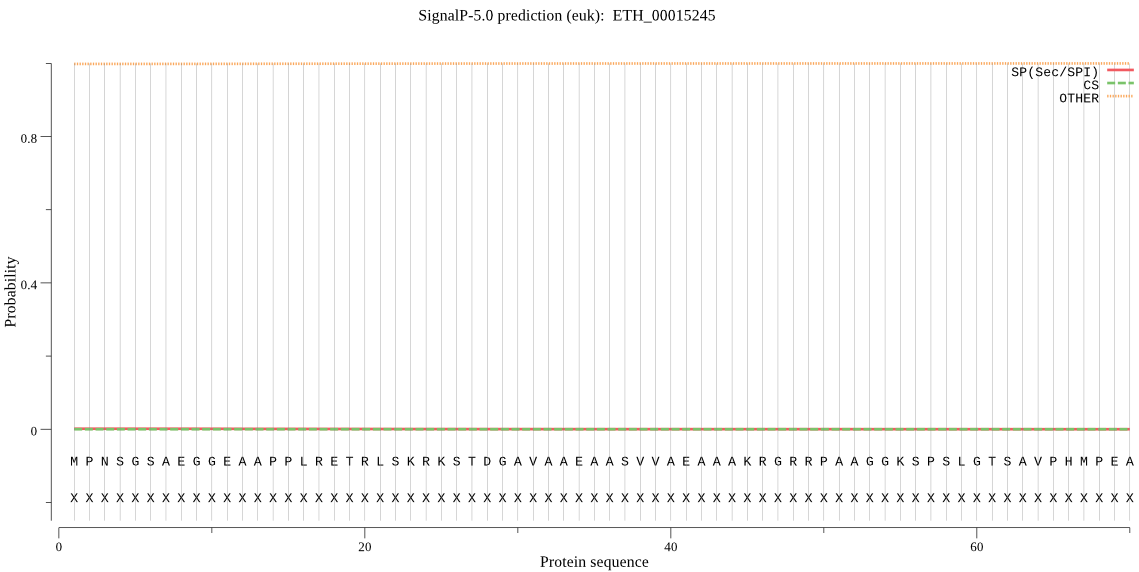
| ETH_00015245 | OTHER | 0.999998 | 0.000001 | 0.000000 |
MPNSGSAEGGEAAPPLRETRLSKRKSTDGAVAAEAASVVAEAAAKRGRRPAAGGKSPSLG TSAVPHMPEAPCNGHVHHPSRAAHLPVHRESDAETKEAIIRSEIPKSVVKVFCTHCEPNY SQPWTTRRQTSSTSTGFVTVDAGGSHCILTNAHSVDNAAVVQVRRRGDHQKHEARVICIG LDCDLAMLQVDDPDFWDGIGPPLEWGPSPSLEDPVTVAGYPLGGDNSSVTQGVVSRTDLQ QYSMGSCWLLAIQIDAAINPGNSGGPALNKEKQCVGIAFQSLKDGDTENIGYIIPSEVVV HFLEDFQRHKKYTGFGDCGFTWQKLENRFMRSALSLKSKQQGVLVKKVDGASYARDVLQR GDIVLAVNGNRVASDGSVSFRNGERILFSWLFAQLFVGDRCSLTILRRGKQLEVSYEVGK VNLLVPATNDLPRPEYLIVGGLVFVPLSEPFLKSEYGDDFESRAPVRLLDRWQHGMQQFP GQQCVILTHVLAHEITVGFEHLHNIQVVAFNGEQVRTLRHLKDLVEASTDEFWRFDLDHE EVVILKAASARSALKSILSRNLIPSHKSEGL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00015245 | 91 S | VHRESDAET | 0.997 | unsp | ETH_00015245 | 91 S | VHRESDAET | 0.997 | unsp | ETH_00015245 | 91 S | VHRESDAET | 0.997 | unsp | ETH_00015245 | 131 S | RRQTSSTST | 0.996 | unsp | ETH_00015245 | 235 S | QGVVSRTDL | 0.995 | unsp | ETH_00015245 | 281 S | IAFQSLKDG | 0.994 | unsp | ETH_00015245 | 379 S | DGSVSFRNG | 0.992 | unsp | ETH_00015245 | 415 S | QLEVSYEVG | 0.991 | unsp | ETH_00015245 | 22 S | ETRLSKRKS | 0.997 | unsp | ETH_00015245 | 26 S | SKRKSTDGA | 0.996 | unsp |