

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
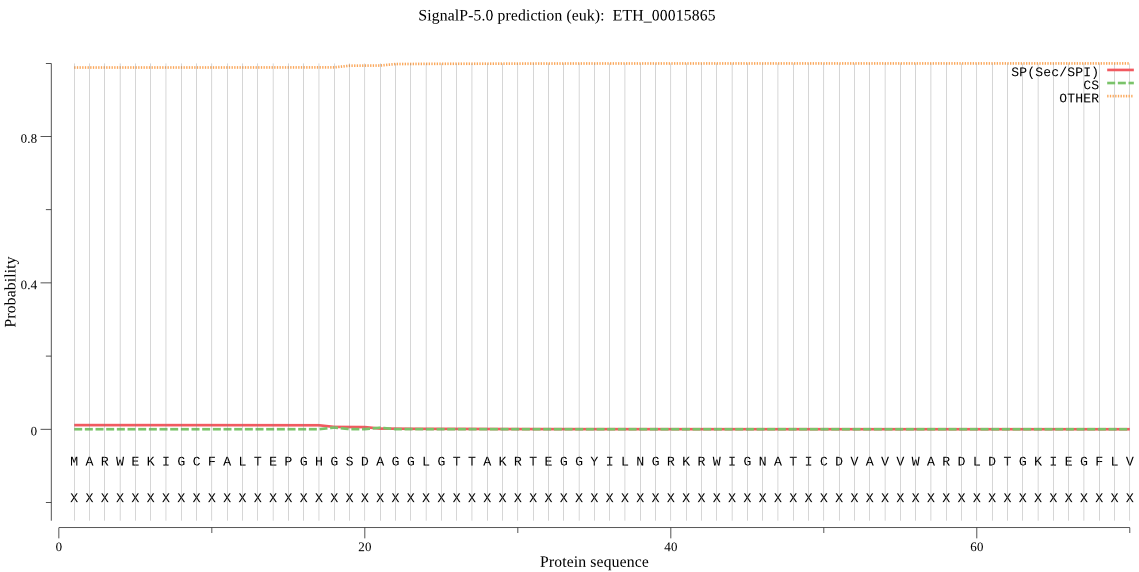
| ETH_00015865 | OTHER | 0.996062 | 0.003274 | 0.000664 |
MARWEKIGCFALTEPGHGSDAGGLGTTAKRTEGGYILNGRKRWIGNATICDVAVVWARDL DTGKIEGFLVEKSFRGFKTAAIKEKACLRSVENADIWLSDCFVPDSHKMKPGGFTGNTKL VLESSRALVAAACSGLLMGAYDAALQYCSSRQQFGRPLTGFQLVQERLMRAIGLAHGCVT LSINMARRMDEGRNSMALVALVKATTSRLAREGTQLCREVMGGNGILLENGAAKALADIE AMYTYEGTYDINMLIAGRAATGINRGEGLPFSYLNALSGRDLSCFHDTSAAAGLKGVQIG ELEYSFGEDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPELHKVISSLQKSFP GSSYDLIRNNCNHFSDALLKSIVGRGIPPHINRASRYGQWLGCLLPQSSGNTSEHNPKTL SSAPSELFTGVFGGQGHRLGGPQGGGIGEYGIGMVKKLIYMANCFKQEAPSIEDEFGAEG EY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00015865 | 362 S | SFPGSSYDL | 0.994 | unsp | ETH_00015865 | 471 S | QEAPSIEDE | 0.998 | unsp |