-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00016620
MLFLAACCCCCCFLIAGGPARAAAAAAAPPGEPGAPREVPDAAAAAAAAAEAAAIAAAAA
AAAAGPAKALQTPAQVPLKVVGSRVALGVQVGGQQLLLGLDSCKEGLRLFASGSSACTSG
RRGSALQQQQQQQPLAAAQRQQQAAAEDAGSPREAPAGEPAAGAAAQQQQQEQQEQQQGD
GKAKQRKQLAKQQQQQQDCYSPDASNSSTWCLNWHEVCTPFSESPFSCRASKQHDPQYAA
RYKQIVDGIFVSEVRLEGYDTVELVPTHVGGPPGGPPGGPPPPGAPREGTAEMSIPSFFS
LRLSKFPVKLVLERDTDFDAYEGLDGVWGIAGPDLCCREASLWNLLLEPAVKGVGLDINL
PPPRAAAAAAAAAAVAAPAAEAPPEPGPPAEVPSFIYFSQEARSIFGEMEWAQRMQTGAA
GVDALMHFTTFDWKLCGEQVGHPLSRSWEAIVDLSSECLVVPPPMWKSLRAWLPLDVEHE
LCKDPEAEEDSLNAHISRPVSLQGEAGEKTYRRTCPLLQRPNSRPLPSLSFSLTQTQSDP
PRLQIPLEKLVIHEEGIGDVLCVVPQPFLSMGADQWRQIRFGTRVLSLFKLVMDRDNWRV
GLQEKHAEPANDVACAARAVCRGDQVYVPYTNRCADPDCSVKILFEMNPETKVCELAWWV
PGAVVAAIAALVTLELYVLHLRSRVVAEACSGRR
- Download Fasta
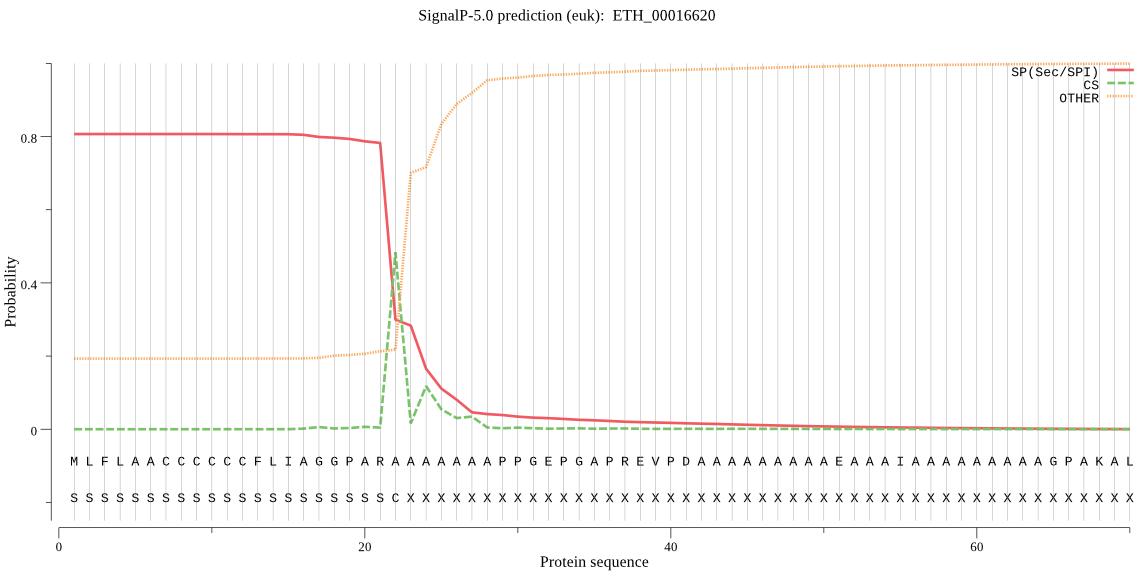
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00016620.fa
Sequence name : ETH_00016620
Sequence length : 694
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.474
CoefTot : 0.543
ChDiff : -16
ZoneTo : 31
KR : 1
DE : 0
CleavSite : 23
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.359 2.553 0.921 0.820
MesoH : 0.431 0.988 0.068 0.389
MuHd_075 : 16.358 8.656 3.226 4.437
MuHd_095 : 14.395 8.924 5.057 3.629
MuHd_100 : 13.255 7.124 4.346 2.707
MuHd_105 : 14.648 6.586 3.462 3.362
Hmax_075 : 12.250 16.700 1.806 5.203
Hmax_095 : 10.600 17.200 8.367 4.462
Hmax_100 : 13.700 14.300 6.472 5.130
Hmax_105 : 14.817 14.817 6.472 4.600
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.8753 0.1247
DFMC : 0.9223 0.0777
-
Fasta :-
>ETH_00016620
ATGCTCTTTCTTGCTGCTTGCTGCTGCTGCTGCTGCTTCCTCATCGCCGGGGGCCCCGCG
CGCGCTGCAGCAGCTGCAGCAGCTCCGCCGGGGGAGCCGGGGGCCCCGCGGGAAGTGCCC
GACGCAGCGGCGGCGGCGGCGGCCGCAGCAGAAGCTGCAGCAATAGCAGCAGCAGCAGCA
GCAGCAGCAGCAGGACCCGCCAAAGCCCTCCAAACCCCAGCCCAAGTGCCCCTTAAAGTT
GTGGGCAGCCGCGTGGCCCTGGGGGTCCAAGTGGGGGGGCAGCAGCTGCTGCTGGGGCTG
GACTCTTGCAAGGAGGGTTTGCGGCTGTTTGCTTCTGGTTCTTCTGCATGCACTTCTGGC
AGGCGCGGCTCCGCGCTGCAGCAGCAGCAGCAGCAGCAGCCCCTTGCTGCGGCGCAACGG
CAGCAGCAAGCTGCTGCGGAGGACGCCGGCAGCCCGCGGGAAGCGCCTGCGGGAGAGCCG
GCAGCGGGTGCAGCGGCGCAGCAGCAGCAGCAGGAGCAGCAGGAGCAGCAGCAGGGGGAC
GGCAAGGCGAAGCAGCGCAAGCAGCTAGCGAAGCAGCAGCAGCAGCAGCAAGACTGCTAC
AGCCCCGATGCCTCAAACAGCAGCACTTGGTGCCTCAACTGGCACGAAGTTTGTACCCCT
TTTTCAGAGTCTCCTTTTAGCTGCCGGGCTTCGAAGCAGCACGACCCCCAATATGCTGCT
CGCTATAAGCAAATTGTCGACGGCATTTTTGTTTCTGAAGTTCGCCTCGAGGGGTACGAC
ACGGTGGAGCTGGTGCCGACCCACGTGGGGGGCCCCCCAGGGGGGCCCCCCGGGGGCCCC
CCCCCACCAGGGGCCCCCCGAGAGGGTACTGCAGAGATGAGTATCCCTTCTTTCTTCTCT
CTCCGCCTCAGCAAGTTTCCGGTGAAGTTGGTGTTGGAAAGAGACACGGACTTCGACGCT
TATGAAGGTCTTGACGGAGTTTGGGGAATTGCAGGGCCGGACCTCTGCTGCCGCGAAGCT
TCGTTGTGGAATTTGCTTCTGGAGCCGGCGGTGAAGGGCGTGGGACTGGACATAAACTTG
CCGCCGCCGCGTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGTTGCTGCGCCTGCAGCA
GAGGCCCCGCCGGAGCCGGGGCCCCCCGCCGAGGTGCCTTCTTTCATTTATTTTTCCCAA
GAGGCCCGCAGCATTTTCGGAGAAATGGAGTGGGCGCAGCGCATGCAAACGGGGGCTGCT
GGAGTGGACGCTTTGATGCATTTCACAACTTTCGACTGGAAACTCTGCGGCGAGCAAGTG
GGACATCCTCTTTCGCGATCTTGGGAAGCAATTGTGGACTTGAGCTCGGAGTGCCTGGTG
GTGCCGCCGCCCATGTGGAAAAGTCTGCGCGCTTGGCTGCCGCTGGACGTGGAACATGAA
CTTTGCAAAGACCCTGAGGCTGAAGAAGACTCTTTGAATGCACACATATCTCGCCCAGTG
TCGTTGCAAGGAGAAGCGGGAGAGAAGACCTACAGACGCACTTGCCCGCTGCTGCAGCGA
CCCAACAGCCGGCCGCTTCCTTCTCTGTCTTTCTCTTTGACTCAAACCCAAAGTGACCCC
CCCAGGCTCCAAATTCCCCTTGAAAAACTTGTCATTCATGAAGAGGGAATAGGAGATGTC
CTCTGCGTCGTGCCGCAGCCCTTCCTTTCCATGGGCGCTGACCAGTGGCGGCAGATCCGA
TTCGGCACCAGAGTTCTCTCCTTATTCAAGTTGGTGATGGATCGCGACAACTGGCGAGTG
GGACTTCAGGAAAAGCACGCGGAGCCGGCCAACGACGTTGCCTGCGCAGCCCGCGCAGTC
TGCAGAGGAGACCAGGTCTATGTGCCCTACACTAACAGGTGTGCGGATCCTGACTGCAGT
GTGAAGATTCTGTTTGAAATGAATCCTGAAACAAAAGTCTGTGAACTGGCGTGGTGGGTT
CCGGGTGCGGTGGTGGCCGCAATTGCAGCTCTTGTCACACTAGAGCTTTACGTGCTTCAC
CTGCGCAGCAGGGTTGTGGCTGAAGCTTGCTCTGGGAGGCGCTGA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00016620 | 151 S | EDAGSPREA | 0.998 | unsp | ETH_00016620 | 227 S | ESPFSCRAS | 0.992 | unsp |


