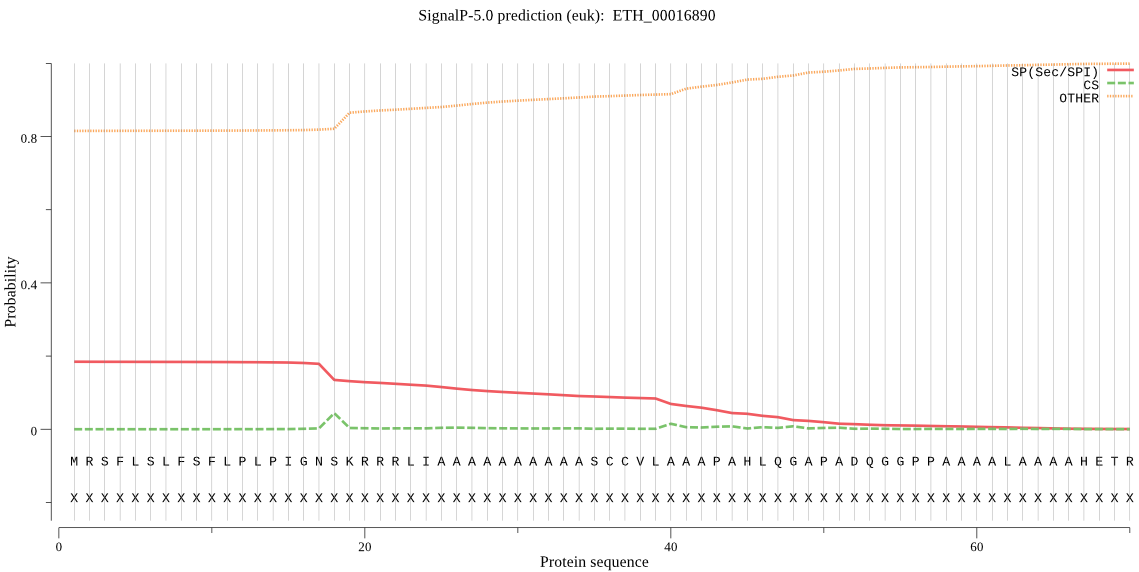
Fasta :-
MRSFLSLFSFLPLPIGNSKRRRLIAAAAAAAAAASCCVLAAAPAHLQGAPADQGGPPAAA
ALAAAAHETRAPSLGAEEALVLGDASSDGGPPNSRDPPDLGAPHDWGAPNGWGAPSDWEG
HNWGAPNELEAPNDFGGSSGAPRDLEDPTGWEPHEIPTPSSAAAAAAAAVSAATESAGAP
SEAGAAATEAAAAEEATAAQRTAAAEQTAAVEAAGDGGSTDAEAAGAAAAAVGAAEAEEP
CPFLSAFFAAQSPPTAAAAAAAAAAAAAAAAAAAGVPLLDAEVPPSSCWVSAEKERMRLA
AAQQTTKLVLAYSLEGCQVAAAAADGAAAAAEEVEVLHGQEARSTAVAALKFKVSQLLRQ
KQLKHTEALLSTDLMRAAAAVDPAAAAAAAAVSPDVSGMETRYLSALEMEIITKLPEGVS
PNEVLSVTPEIPCVQRGFRDSSKFIEEPDPPPRQQQQQQQQQQQQQQQQEAQGESFGAAR
RLQQGPGLLELSRLVIASAAASAAPPRSAAAAAAAAAAAAAGEAVPPNDPFFADQWNLQE
AAPFGINAEKVWRLWTGSSRPMVIAVIDSGCDLNHPELQRSKWTNPAEVCGDGVDNDNNG
FVDDCHGWDFVGNSGELRPDASGHGTGAAGVVGSAANNAVGLAGVCWGCRIMCLKFIGNG
QGTVANQVQAIDYAVKMGAWISNNSYGGYGKADLEFEAIQRAQRAGHLFISSAGNHNLNT
DMPANDHTPSTYPLSNILSVGALTPEGRRAAFSNYGVRTVHIFAPGTGIPTTDGASGFST
VNGTSFACPHVAGAAGLIWSAFPNLTYLEVKRALMESCRQTKALKGLAACGGSLDVYGAL
VLAAHISSGIPLDKSALSRATAQLPPPPEAAVAAAETKGATEAPTQQQQQQPQQQQQLQQ
KQQKFEPRNPSPAAAALTGIQNLPRDPLTLLSSVMFAPLPLVDDNSSSSNSSSNSNSNSN
SNSSNSNSSSGKSRPGRSSLLSLLPLGPREEAALDEDSLQQQQQQLQPTTSNAAAEPAKH
TEAFNLTMFSLQQQLLHPQQQLLQPQQASQQQQEQQQLQHPQNQLHQMPKQQQLLQQQPL
KQTQLQQPQLHQTQLQQQSLEQQQLQQPQLQQQQLQQQLHQQYLQQLQLQQLPQLQQQQQ
QLQQQQLQQQQLQQQQLQQQQLQQQIPQQQLQQQQLSQQQQLQQQQMQQPQLQQLLFRQP
QPLQQLLQQQQLMAREPQLLQIEALSNEKLNEGLQQQQQQPMLPQQQPLLQQQQPLLQQQ
LLPQQQLVSQQQLLPSQKLLLQQQKLLQQQQLLQQQLLQQQQQIAAGVSIESASQEQQQL
LQQHQLVKQQLQQQQQQLAALLSIDSASQEQQPMLESDVLQENLLQQQRLLQLQQQEQQK
KLLQMQQQQLQLQQQLQQQQLLQQQQQKASHMKALRHGSQAQQQPPQKQQQQQQKSQQQQ
QLSLQQQLLQLWKQPQELLFSGSLPSSVLRLLPFAETPAQEGLRKGSSSSSSSSKTLLQL
V