-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
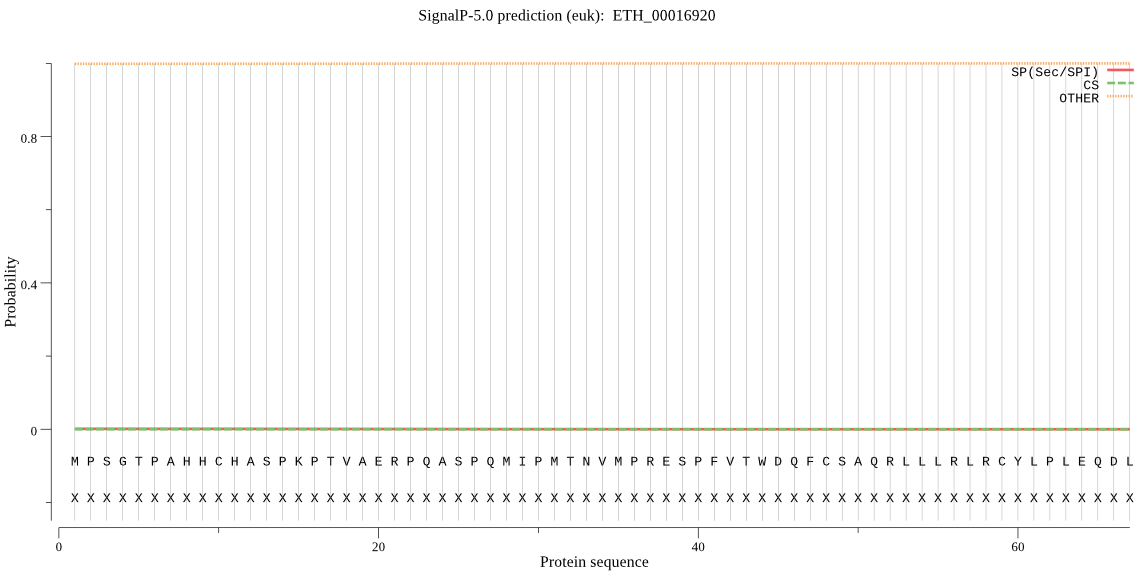
Fasta :-
>ETH_00016920
MPSGTPAHHCHASPKPTVAERPQASPQMIPMTNVMPRESPFVTWDQFCSAQRLLLRLRCY
LPLEQDL
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00016920.fa
Sequence name : ETH_00016920
Sequence length : 67
VALUES OF COMPUTED PARAMETERS
Coef20 : 2.902
CoefTot : -2.611
ChDiff : 1
ZoneTo : 37
KR : 3
DE : 1
CleavSite : 39
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 0.071 0.688 -0.175 0.211
MesoH : -2.795 -2.197 -2.335 -1.883
MuHd_075 : 24.631 16.977 6.388 5.056
MuHd_095 : 15.497 10.662 6.643 3.117
MuHd_100 : 16.687 12.661 6.022 2.516
MuHd_105 : 30.029 17.129 7.912 4.358
Hmax_075 : 5.717 6.300 -0.993 2.707
Hmax_095 : 0.800 1.600 1.690 0.920
Hmax_100 : 2.100 3.000 1.690 1.980
Hmax_105 : 11.300 8.750 0.139 2.690
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9811 0.0189
DFMC : 0.9734 0.0266
-
Fasta :-
>ETH_00016920
ATGCCGAGCGGGACGCCGGCCCATCATTGCCATGCTAGTCCGAAGCCCACCGTGGCAGAG
CGGCCGCAGGCGTCCCCGCAGATGATACCTATGACCAATGTTATGCCACGGGAGAGTCCC
TTCGTCACGTGGGACCAATTCTGTAGTGCACAGCGCCTACTGCTGCGACTACGCTGCTAC
CTGCCCCTCGAGCAGGACCTATGA
- Download Fasta


