

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
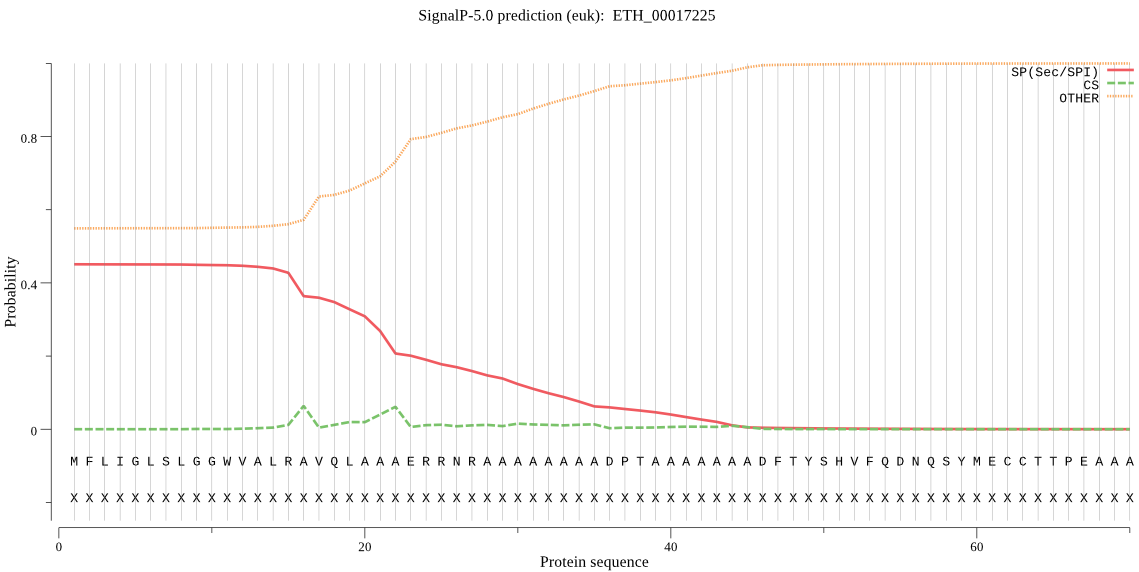
| ETH_00017225 | OTHER | 0.439972 | 0.232448 | 0.327580 |
MFLIGLSLGGWVALRAVQLAAAERRNRAAAAAAAADPTAAAAAAADFTYSHVFQDNQSYM ECCTTPEAAAAAAATDADSAAAAGAAAAAAEAPLGLRGLVLLSPMLQLSRRRYLQRSRLL QFAAAAFAAVRPHAAVVPPRKSRRRHPHLHAYYSSDPLTYKEGVPAGIAVPLLWEMEAAT TPQELLLLQQQDAAAVLMLHTVHDPLCDISGPVSCFSELAGIRDRQLLAIEGPPLPAAAA GAQQQQQQQDASSSSGDPQQHVHPKLRKLHPYFHQHLPDPPSPQPHQPQQQQRQQQQQQE VAASVEEIERRMQQLSQTGEQQPKVFIAKGVDVLHDLPNEPGQQHLFRLLATWITCRCP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00017225 | 142 S | PPRKSRRRH | 0.997 | unsp | ETH_00017225 | 304 S | EVAASVEEI | 0.996 | unsp |