-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
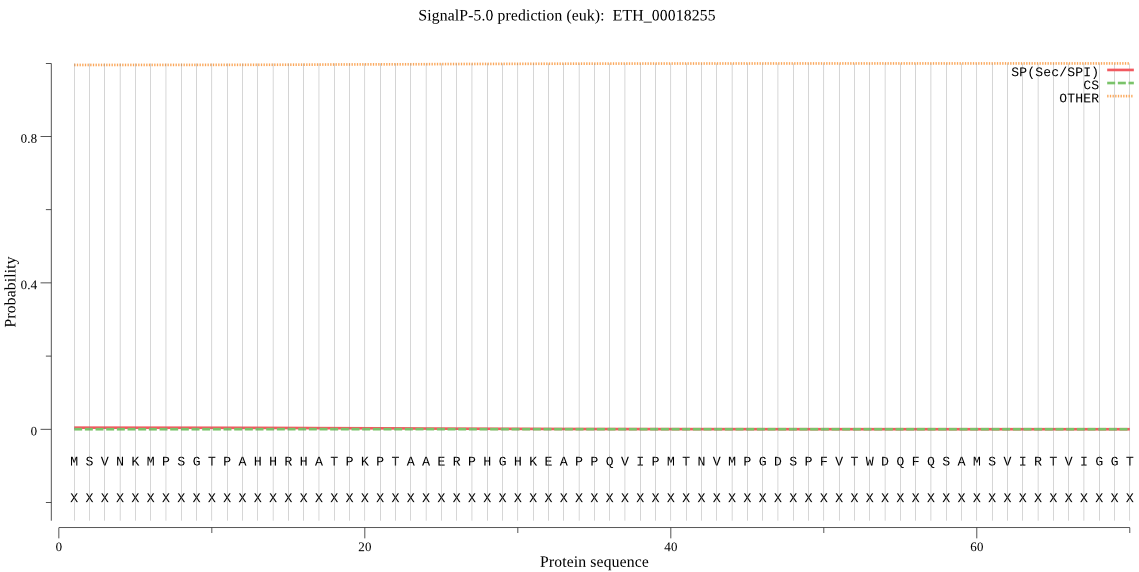
Fasta :-
>ETH_00018255
MSVNKMPSGTPAHHRHATPKPTAAERPHGHKEAPPQVIPMTNVMPGDSPFVTWDQFQSAM
SVIRTVIGGTMESGATIPILPQDGSTMQAFLLRIERRYTQMGLEPRVWGNAFIDHLVGPT
LTYWMYLRRTIDLSNWARVQRRLLERFDKTMSQSQLLTELAKVRWNGNPKEYTDRFAAVA
ARGASNAQQNALCHSSESLRGTAEPALGKASMRCKPCNAHRSY
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00018255.fa
Sequence name : ETH_00018255
Sequence length : 223
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.257
CoefTot : -1.416
ChDiff : 10
ZoneTo : 24
KR : 3
DE : 0
CleavSite : 17
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.076 0.847 0.084 0.517
MesoH : -0.252 0.165 -0.345 0.207
MuHd_075 : 30.914 18.112 8.946 6.693
MuHd_095 : 23.537 11.950 6.612 4.160
MuHd_100 : 23.042 12.625 5.017 4.834
MuHd_105 : 22.030 12.077 4.039 5.290
Hmax_075 : 3.733 2.567 -1.755 2.543
Hmax_095 : 6.800 1.700 -1.682 1.980
Hmax_100 : 7.400 2.800 -1.682 2.210
Hmax_105 : 6.533 2.800 -1.866 2.368
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.4823 0.5177
DFMC : 0.4511 0.5489
This protein is probably imported in mitochondria.
f(Ser) = 0.0833 f(Arg) = 0.0417 CMi = 0.64935
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>ETH_00018255
ATGTCAGTCAACAAAATGCCGAGCGGGACGCCAGCCCATCATCGCCATGCGACTCCGAAG
CCCACCGCGGCAGAGCGGCCGCATGGACATAAGGAGGCGCCCCCGCAGGTGATACCTATG
ACCAATGTTATGCCAGGGGACAGTCCCTTCGTCACCTGGGACCAATTCCAGTCCGCAATG
AGTGTGATAAGAACTGTTATAGGGGGGACCATGGAATCCGGCGCAACGATTCCCATACTA
CCACAAGACGGCAGCACTATGCAGGCCTTTCTGCTGCGCATCGAGCGCCGGTACACCCAG
ATGGGATTGGAACCACGTGTGTGGGGAAATGCATTTATTGACCACCTTGTGGGCCCGACT
CTAACGTATTGGATGTATCTACGGCGAACTATCGACCTAAGCAACTGGGCGAGAGTACAG
CGCAGGCTTTTGGAGCGGTTTGACAAAACAATGTCGCAGAGTCAATTGTTAACGGAGTTG
GCTAAAGTACGGTGGAACGGTAACCCGAAGGAGTACACGGACCGGTTTGCGGCTGTAGCG
GCGCGTGGGGCAAGCAACGCGCAACAAAACGCGCTTTGTCATTCATCCGAGAGCCTTAGG
GGCACCGCCGAACCGGCTTTGGGCAAAGCCAGTATGAGATGCAAACCGTGCAATGCCCAT
CGCTCGTATTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00018255 | 18 T | HRHATPKPT | 0.996 | unsp | ETH_00018255 | 61 S | QSAMSVIRT | 0.996 | unsp |


