

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00019755 | SP | 0.001900 | 0.998094 | 0.000006 | CS pos: 42-43. VAA-DL. Pr: 0.7876 |
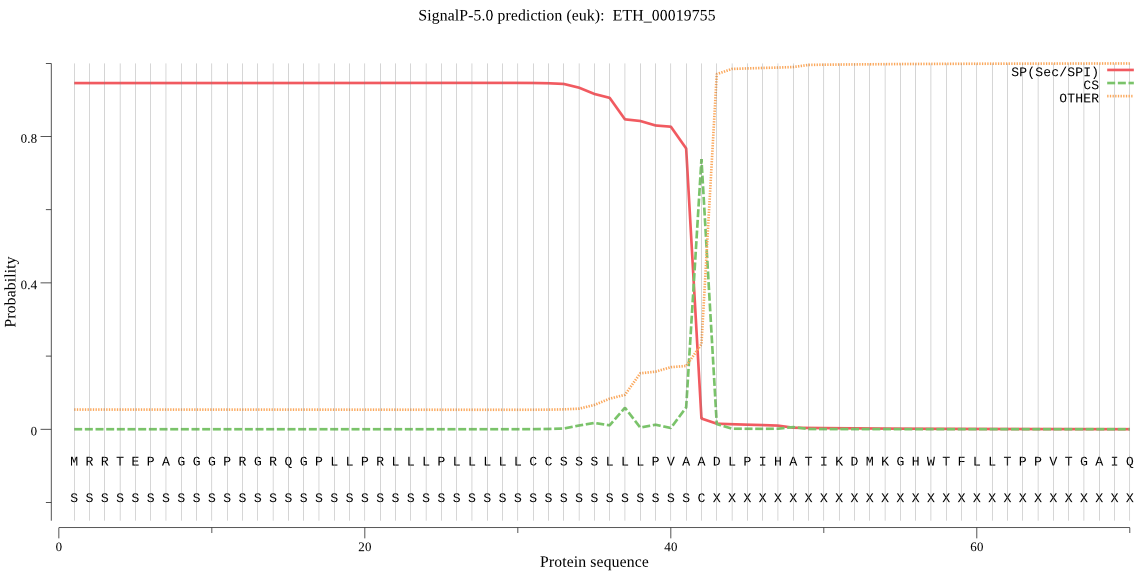
MRRTEPAGGGPRGRQGPLLPRLLLPLLLLLCCSSSLLLPVAADLPIHATIKDMKGHWTFL LTPPVTGAIQSCGSGAPNKNTENLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVY DTKDNLHRNEWKMLAVKDKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVKQNCPEA KDGDLEDSNGETVCYSTDPSKTQLGWYSSTAADGSVASGCFFGEKEEKDSAAAAAAFVVL HQRAAAAQQQEAAAAAAGGAQELSYFLSEDFVLQHNSKKSSWIAAVNSSFANQNKNFLNS LLKQFQSSPRLLGSSEGGPVFLEEAAEDAASTQAFACPCKEGEKALDTRSKSAAAAAAVL SPVSLAQEESKAKTKTVPVKAHSQQQQQQQEEQQQQQLQQMQQQQEAAVKLSPQSVLSCS FYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEKDFLKSEKDFLK GEINGSGEDSSSNAAFLLSGEATGTCHQGENRWFAKGYGYVGGCYECLSCSAEQKIMKEI MTNGPVAAALDAPPSLFAYSSGVYTTRGAPHARVCDLPGAPSFLPTSSSSSSSSSGSALS GWEYTNHAVTIVGWGETGSGSGSDPQKYWIVRNTWGPQWGLKGFFLLERGNNSGGIENQT SFIDPDLTRGKGLALASASNQLAS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00019755 | 591 S | SSSSSSSSS | 0.993 | unsp | ETH_00019755 | 591 S | SSSSSSSSS | 0.993 | unsp | ETH_00019755 | 591 S | SSSSSSSSS | 0.993 | unsp | ETH_00019755 | 592 S | SSSSSSSSG | 0.99 | unsp | ETH_00019755 | 661 S | ENQTSFIDP | 0.995 | unsp | ETH_00019755 | 474 S | DFLKSEKDF | 0.997 | unsp | ETH_00019755 | 486 S | EINGSGEDS | 0.996 | unsp |