

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00019930 | OTHER | 0.994114 | 0.001287 | 0.004599 |
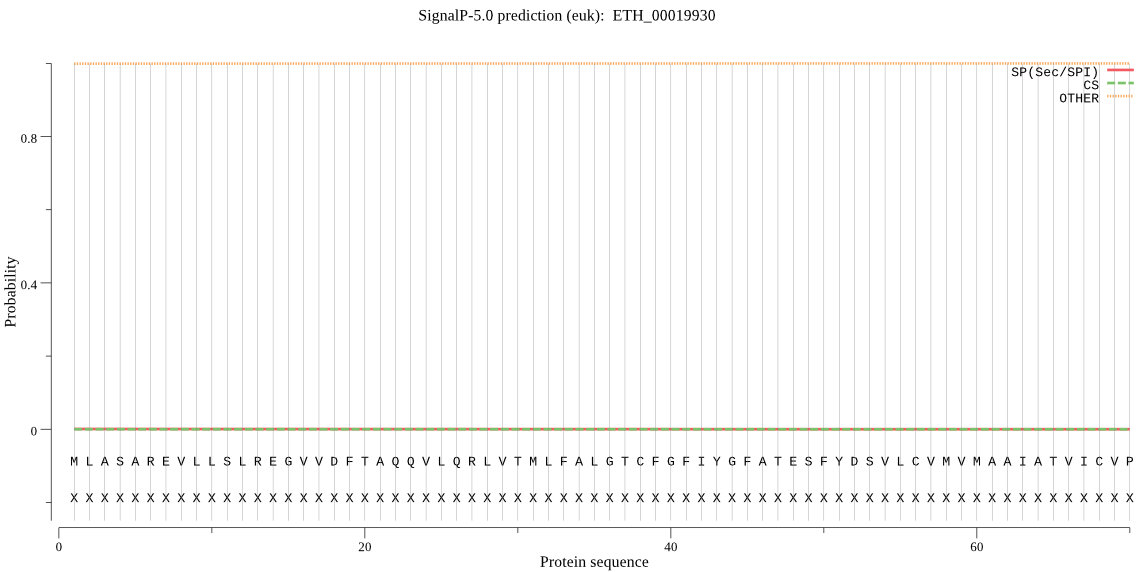
MLASAREVLLSLREGVVDFTAQQVLQRLVTMLFALGTCFGFIYGFATESFYDSVLCVMVM AAIATVICVPSWPLYKRNTIDWAPHDPERIAKLYEEEMAQARSLKEAVSALPSKKKGQVN PKKRS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00019930 | 11 S | EVLLSLREG | 0.995 | unsp | ETH_00019930 | 103 S | AQARSLKEA | 0.996 | unsp |