

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00020065 | SP | 0.029646 | 0.969313 | 0.001041 | CS pos: 33-34. GLA-QD. Pr: 0.5814 |
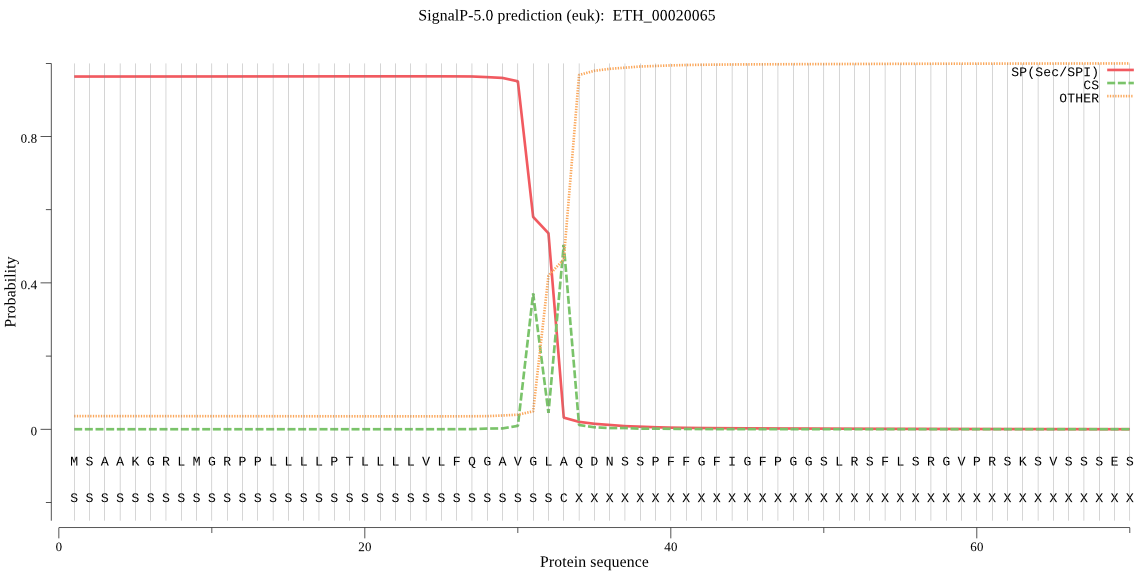
MSAAKGRLMGRPPLLLLPTLLLLVLFQGAVGLAQDNSSPFFGFIGFPGGSLRSFLSRGVP RSKSVSSSESSSSSRREVGASRFTSYEGPLDSSLEKGRGGPGAHASWYGCMWQQVGAEGA AGVGPLRAGDDGLVLDASDYTEKAWEAMGALGSIADKHESAYVEAEMLLLALLQDGPEGL AHRVLSRAGVSVEKLTEEVERHLERQPRMALGFGDQKVLGRGLQQVLASAQRFKREFRDE YLSVEHLLLGLAAEDAKFLRPALQRQGVSFNKIKEAVVEVRGKKRVNTKNPEMAYQALER YSKDLTAEARAGKLDPVVGRDDEIRRVIQILSRRTKNNPILLGDPGVGKTAIVEGLAQRI VSGDVPDSLKGRRVLALDMGALVAGSKYRGEFEERLKTVLKDVQDAEGDVVLFIDEIHTV VGAGASSEGAMDAGNLLKPLLARGELRCIGATTTQEYRQHIEKDKALERRFQRVLIDEPQ VEATVSILRGLKERYEVHHGVRILDSALVEAANLASRYITDRFLPDKAIDLVDEAAARLK IQVSSKPIQLDRLDRLLLQLEMERISILGDSKARALDEQEKLRLRAVESQIERLQAQQAT LTDAWNKEKSQVDAIRAIKERIDVVKVEVERAEREFDLNRAAELRFETLPDLERQLQEAE AQYKAAHAEGRRLLRDEVTADDIAGVVAVWTGIPLTRLKESEREKLLNLKDRLHERIIAQ DKAVSAVAEAIQRSRAGLNDPNRPIASLFFLGPTGVGKTELCRALAEALFASEDAIVRIN MSEYMERHSVSRLIGAPPGYVGFEQGGQLTDAVRKKPYSVILFDEMEKAHPEIFNVLLQL LDEGRLADSKGNVVNFRNCVVIFTSNIGADALLESAGDPSKKQAVEKKVMQTVRDTLRPE FYNRIDEFVVFDALTKQQIKEVVRLEMERVADRLLEKKIKLKVDESALVHLADTGYDPAY GARPLKRLIQRLVETRIAQLLLQGSLQELDTVTVKSENNTLLFSVSSAATGETTDYPASP APTPASPSSAATEQQATLAAPPAAAAQVPAIAPENAAATDTRAQVVTQEPLVPTS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00020065 | 67 S | KSVSSSESS | 0.995 | unsp | ETH_00020065 | 67 S | KSVSSSESS | 0.995 | unsp | ETH_00020065 | 67 S | KSVSSSESS | 0.995 | unsp | ETH_00020065 | 73 S | ESSSSSRRE | 0.996 | unsp | ETH_00020065 | 74 S | SSSSSRREV | 0.995 | unsp | ETH_00020065 | 85 S | SRFTSYEGP | 0.994 | unsp | ETH_00020065 | 368 S | DVPDSLKGR | 0.992 | unsp | ETH_00020065 | 701 S | RLKESEREK | 0.996 | unsp | ETH_00020065 | 789 S | MERHSVSRL | 0.996 | unsp | ETH_00020065 | 64 S | PRSKSVSSS | 0.994 | unsp | ETH_00020065 | 66 S | SKSVSSSES | 0.997 | unsp |