

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
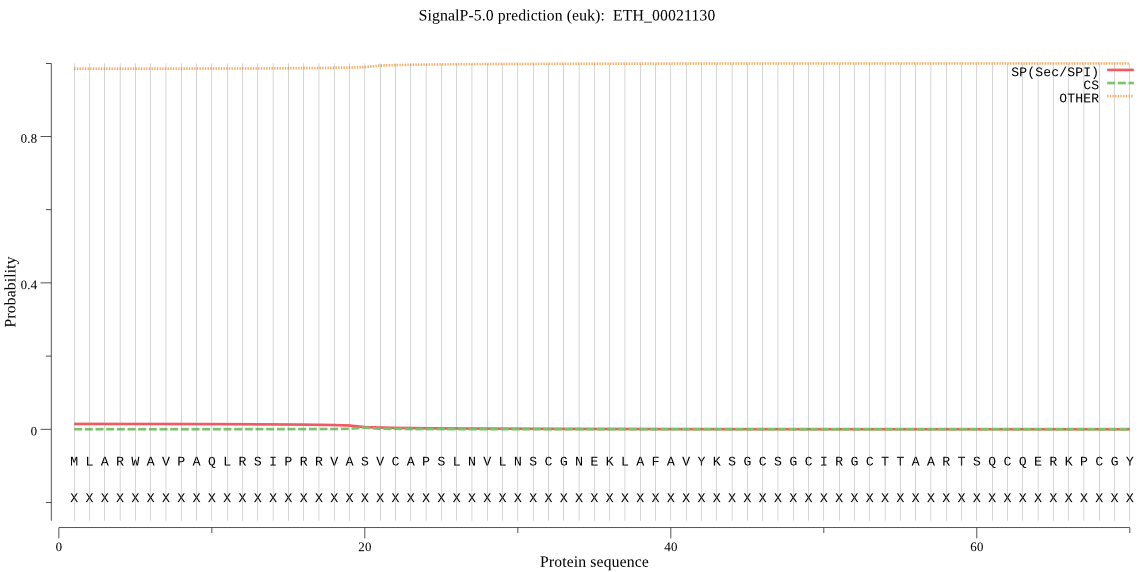
| ETH_00021130 | mTP | 0.320996 | 0.003943 | 0.675062 | CS pos: 59-60. ART-SQ. Pr: 0.0814 |
MLARWAVPAQLRSIPRRVASVCAPSLNVLNSCGNEKLAFAVYKSGCSGCIRGCTTAARTS QCQERKPCGYCSLWPKEARRGRHILALQSSAANQLQDGPRSRWRGSQGAFQPVPQPAFEN EPPLQIETGKSVASLRDMSRSPQALVKYLDNYIVGQTQAKRAVAVALRQRWRRRQVADPE MRADITPKNILLIGPTGVGKTEIARRLAKHLDAPFVKVEATKYTEVGFHGRDVDQIIKDL VEVAVKQKRTILESQLRFAAKEKAEAKVLEALAGKMPVSWLRGEDTEGCS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00021130 | 139 S | LRDMSRSPQ | 0.991 | unsp | ETH_00021130 | 139 S | LRDMSRSPQ | 0.991 | unsp | ETH_00021130 | 139 S | LRDMSRSPQ | 0.991 | unsp | ETH_00021130 | 60 S | AARTSQCQE | 0.995 | unsp | ETH_00021130 | 134 S | KSVASLRDM | 0.996 | unsp |