

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00021535 | OTHER | 0.999446 | 0.000280 | 0.000274 |
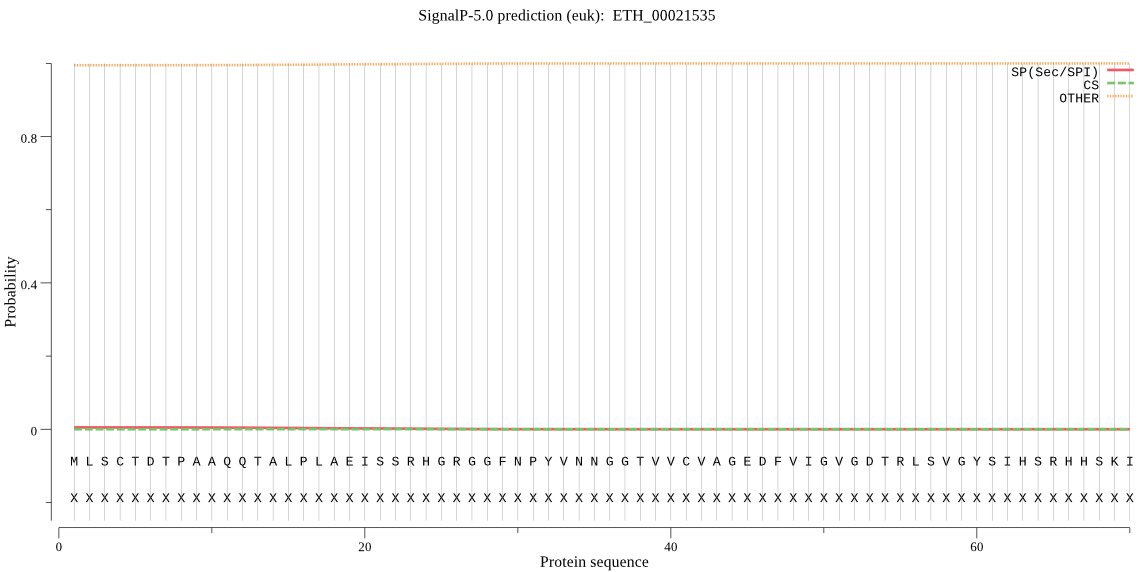
MLSCTDTPAAQQTALPLAEISSRHGRGGFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGY SIHSRHHSKITQLTSRCCIASSGMQADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLS SLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQM PLVTALLDNQGKKKKRNEERKKRERKEK