

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00022120 | OTHER | 0.999975 | 0.000025 | 0.000000 |
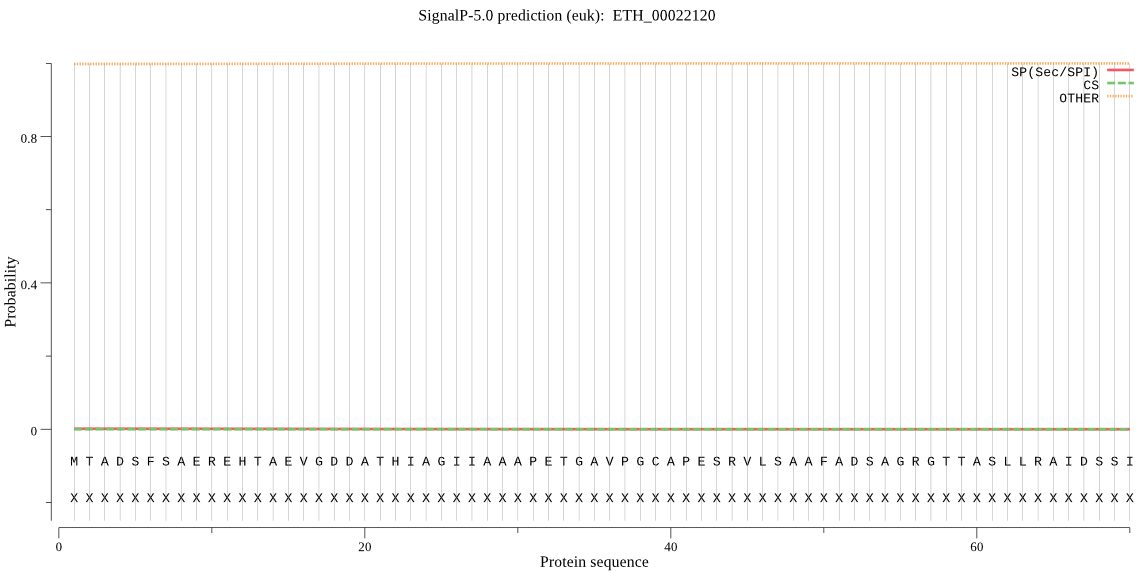
MTADSFSAEREHTAEVGDDATHIAGIIAAAPETGAVPGCAPESRVLSAAFADSAGRGTTA SLLRAIDSSIEQGVSVLLNSWQATAAAAASSSSSSSSSSIYRALSFAAKANGGKGILIIN AAGGSQKEIDEGTTLYPISLGIDNTINVAALSPSGSLASYSNYGASVHLAAPGEKILSSA TNNGHKIRAATSAAAAFVAAAAALVFGVFTEFRSAAAAAEVKELLKATAAAKETLQQHTQ WGAAVDAAAAVMAAQLGGAFIQVDCMDRKFLLKPRESRVVSVYVRAFIPGEFKAFLNVGV RAAKQFSGEAKEGQIPIWFNSFKFISAEEIEENDLTSLEAETAFEDLLLQQQQQQGDPAA AAEDQQQQQDSENYGEGEKDDLEDSGEINSQKEFENENEKFCKVQDGFVQVISVRDLLGE EEPEEGFGYTGAIVAGASISFLAIGGVFYLVQNK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00022120 | 390 S | GEINSQKEF | 0.992 | unsp | ETH_00022120 | 390 S | GEINSQKEF | 0.992 | unsp | ETH_00022120 | 390 S | GEINSQKEF | 0.992 | unsp | ETH_00022120 | 7 S | ADSFSAERE | 0.993 | unsp | ETH_00022120 | 99 S | SSSSSIYRA | 0.993 | unsp |