

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
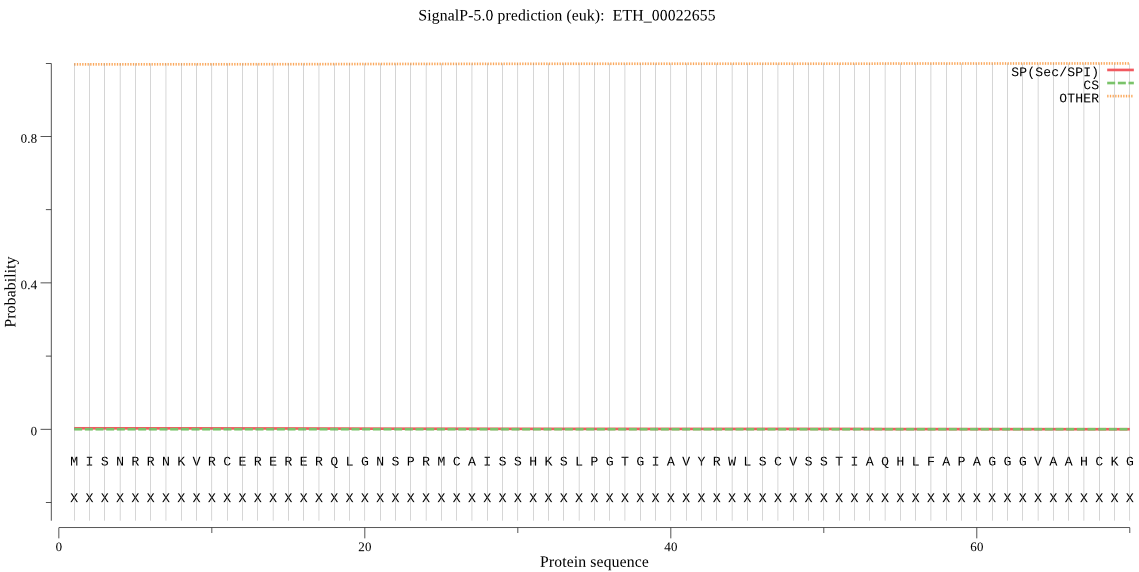
| ETH_00022655 | OTHER | 0.996410 | 0.000269 | 0.003322 |
MISNRRNKVRCERERERQLGNSPRMCAISSHKSLPGTGIAVYRWLSCVSSTIAQHLFAPA GGGVAAHCKGIRGQCVEQEEQRPHETLAPEQAFEMMKPGVVRVYAILPELASACQQEGCM ASPGRNPHVVERFHFLGSGFFFDDRGHVVTAAHVVLRLGDSKEESALNSTADFKQDQGLQ GDFVPYRLAIKDSVGRIHPANLCGLDTVTDVAVLRVEGITNDNSPQTFCKKFDGARPLMG EIVATYAATEHAEESIGVAGRVLQPRQTFKSLDSSGCLGFLQLQLLTLPGMSGAPVCNMS GHVVGMLVKKFDMCGLALPSAVVRRVAEALRDTGKFTSPSIGLLVKEEVPCVASSHQKLP RRPTLTVCGVTPGEAADVAGIQEGDRIISVQGERMDSVVQLRELVVLHGGGSMDVTVRRG TEWTIDCDDFLLKVGRLQENATPAE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| ETH_00022655 | 161 S | RLGDSKEES | 0.994 | unsp |