

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00024440 | OTHER | 0.993335 | 0.002059 | 0.004605 |
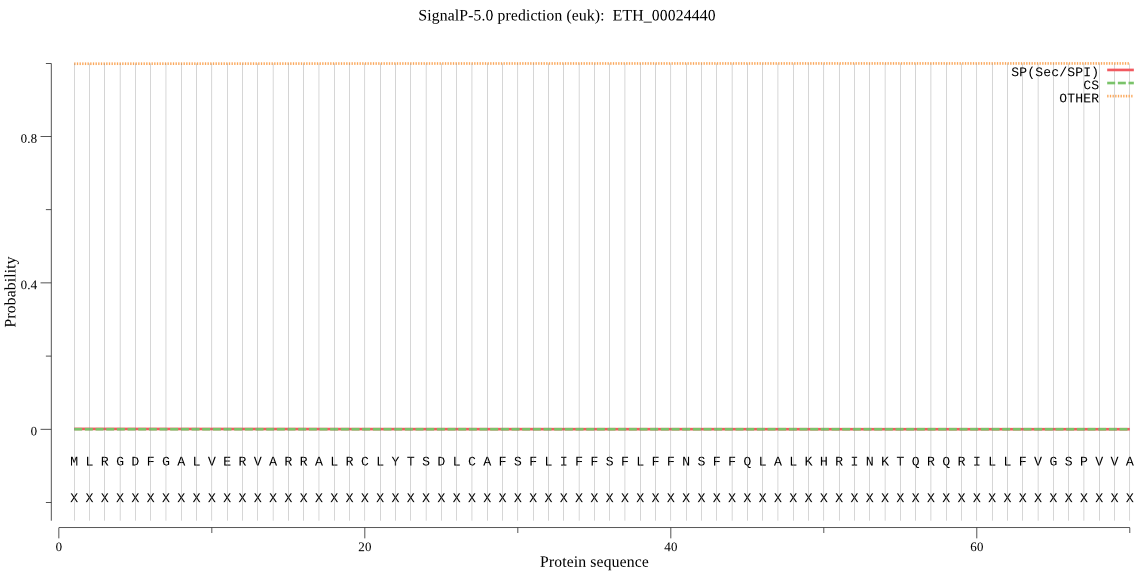
MLRGDFGALVERVARRALRCLYTSDLCAFSFLIFFSFLFFNSFFQLALKHRINKTQRQRI LLFVGSPVVASEKQLVQLGRQLKKNNIALDIVCLANVEENRAKLTAMHAAVNSNDTSRFV EYPAGSSSLLSDVMVSAMLLHDPEAAAAAAAAGGDSAVNEFGVDPNVDPDLYMVLRMSLE EARQQQQQQQGAAAGAAAATAPAGQPAAAAGSAELPPGAAQIEGLEDMDEELRQAVLLSL QDYCGDASSTEATQQQQQQQQQEQQQGEGQESQGAANGAGAAAAAETEAAAAAAPNNANE QPPGDAAEKEPKGSS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| ETH_00024440 | 178 S | VLRMSLEEA | 0.998 | unsp |