

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00025145 | OTHER | 0.765601 | 0.226085 | 0.008314 |
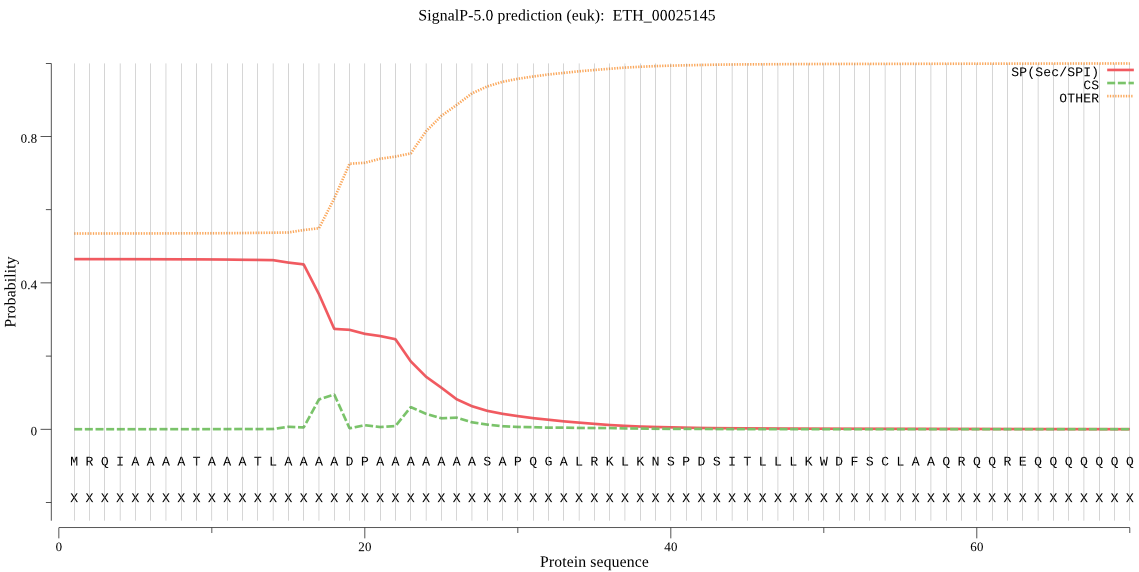
MRQIAAAATAAATLAAAADPAAAAAAASAPQGALRKLKNSPDSITLLLKWDFSCLAAQRQ QREQQQQQQQQQPVEVYLQLDRDVTRAAAAAEAGKGQVLFTRGVFLRDKAQKQQQQQQQQ QQKQQQQKQKQKQQQQQQQQQQFDDRVFGLQFLSQLLGHNIDLEAEAAELQQELQQELQQ EQQQEQQQELLPQRQQADGAAEQQKSPCVSSPSLCQELLLQKLLQQQEGLTVSKYLSSFD VDVVTFSRLKEFMRESGLQSEEELLQLLKETHSSSCLSQVSLDMKGTLTGSVPAAAAAAT AARGSSPRAAAAAANVEDPYFPLQWHLQAAEGLYNLGADKAWQQMQSAQQQQPQQQQVVV ALLDTGCGPHEDINNILWINSKENCSDGVDHDNNGYVNDCKGWDFVDDTNDITDTYGHGT AVAALMAASFDKKGGRGKP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00025145 | 260 S | SGLQSEEEL | 0.991 | unsp | ETH_00025145 | 306 S | ARGSSPRAA | 0.996 | unsp |