

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00026815 | SP | 0.010673 | 0.988536 | 0.000791 | CS pos: 24-25. GSG-RL. Pr: 0.6530 |
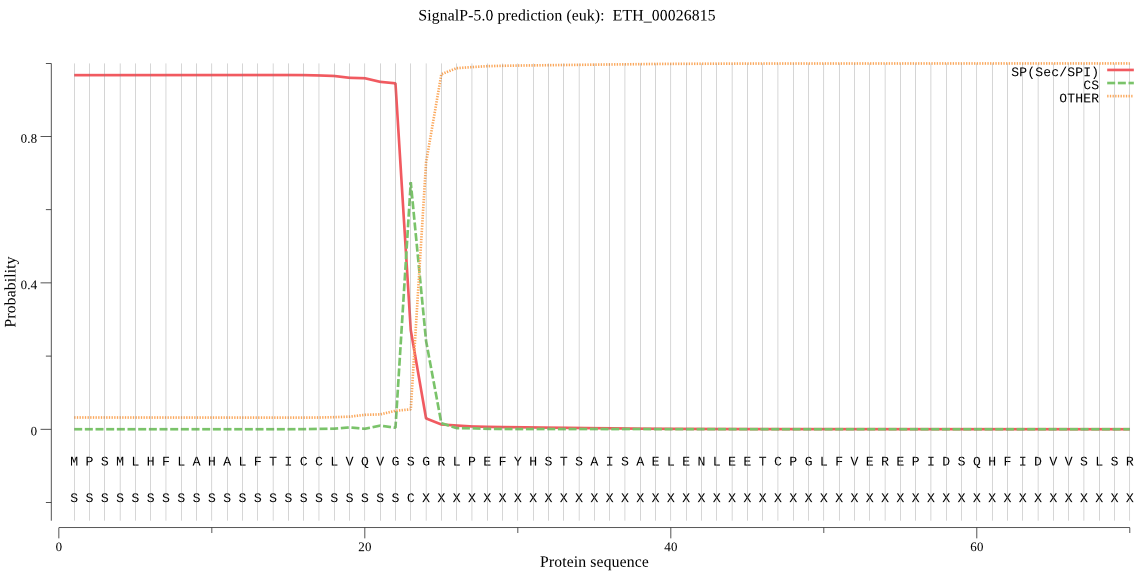
MPSMLHFLAHALFTICCLVQVGSGRLPEFYHSTSAISAELENLEETCPGLFVEREPIDSQ HFIDVVSLSRSENPVSHFFLLAGEHARELITAESALHFLKVLCGEEPALRKKAEAALADT SFTVIVNGNPISRAKVENGDFCVS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| ETH_00026815 | 69 S | VVSLSRSEN | 0.995 | unsp |