-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs: GO:0005509
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00028250
MQEETQRFGGHLFFSLGFWWLLASRVCSTCLAFTATGYIPLATTAPTEAKLPDAAEALPP
VFVQEPSRGALPVCDCTGLHHGTVHDAACSIGFSDEATDFCFIPPSYCATVESGEMLTVN
GSRLEEYSILLVDWGKTKESADYYEKALPRASTPTCRCWPFNGVMVAPCEQQWPPQVRAE
GTGGSNTSAGDLSAASESILAGGVLSRRQSALKKSFVSGGDAPDAAIHRNLFHTPIQATG
LQRLSKWARHLAVWRQAITGDSHSSKDVTAPSVNTTCHKGCASCLQPGRDRCLTCFNQLP
YPYRPLPAGSHPPNPNKLPSDIYGSNAYKKQYHAHIAAHRDGSGYCIPLAFGGDALAAAH
SSCLASDSSCVKKNVRPLEQPEGGFLELNNGSHCHASCAACRADCCRGKADNCLACTGPS
RIFHSLYGDGTGRCIDLRLPDDQVQTTNLVEKGQDEGMMYMQQLANFEGEETGSFSAVLQ
SIISIAFSSPAAPDTGKPKVAASLDKSRNRKGPSFLPRRSICDSLPQRIGDMPTRAWSQL
MFVHADNNLEESALLDLEEMLHPWRGEVVKRYARAAMRGRGTPPLPVGGSALSGSTAQEA
LEDLYLVVLIDRSKQTTSTDMGTVHACPELEYSSLGQGVRQLPGTSEVELNAQMAFELLR
IHLQDGRREWLLLRSLGEVDMNDPGVLRTAVTRLLDIFPSRHYAVILWNHGSAWAGFGDD
ESNPNNQPMSIHDIALGLKKSMDPTSLTRKPPPPYRTATAFEYGARFVTSYGLHPPSSNA
LTLALIEVRAFNEFKQKFEELLEGLYSCGGSNITRRVRRALSQTFSVRGCRMLGLCSCYD
LNDFLANLLQQFASEENDLGRISMQSLGHDDELSWDDSNSEMRDITYRRLQHTNGQLYAI
NALGAAQRALTDKVVAARVFFKRMVVAEVGSREPGRYGGLSIYFPDPNMAVNCKTHRSAA
SWARRYTSTIRTKFSSFVSSVLSNRKGSVCYAPWGGTGDAAQDATGTIPQHAAEAPQWFG
ILCSRVLKSYLDKHLGAAVGISAVLPATVASAMMFRGFAASVANNGKPEIIITSTAQATM
TDAEVKSSTVPLTSSGMSEEAQRLHARSKANLPGSRAHDARAEPSDTAAALREFTVVQGW
WDTQVWILKQQLRLDDSLKDPRPRRQTHNGSAKDADVEAIVVAIQDGPASSRFGAGRASL
SCCLLTVLQGTTFSFPFLFFKDALAAECLEDSLLHTEALSAPKSPQNSFESRSTAGQAKA
RMLPADLAGFKKRSLQELSSSPVEDMNKQEASVKRHILSSVVLPLATPSEDFQIVHDVRA
NSQEQASSSSLQASSSDLRGSQKPKFDAELQEKDDDVSKVPQPRQLSSVRAGAASWLLHE
RLGREPQRHVDKKRQCGARAYLLAEWDENRRDSSELVLYVVEDEQATEWPRSKGGLLRPI
EHRLRRTTVFQQYFEQAPSSIPNARGGSPRNKGTSQSGISTNAAYQPRSSATGHQTGFSS
GHGDARFSESLSFTRAAGSQHEAFWGQQADQGSEENVAAVVLEESLGEATFFWNVESEVD
KLVLQPRSISSLLRERRSKMNVEGSQVLENTERPVVGFIAQDLFDQRAVAASAVPLTERQ
SPSFPQSLWESIEQTFGFGDYAQETLQASNALPSSSTCQPYWLGDGICDEICDTAAYEFD
GGDCVHLNAAAEDLLEQQEETPLVATIWEGPRGRYVCRCPKGYTGDGTSCEDIDECKVFE
EAVCHAKAICINTKGSYRPSNQSYSGSCIDIDECSSGLASCASPSIATCVNTSGGYFCSC
VDGYEGDGRQCVASPSTAEAEISIAMDYHKTVAAAGGIQLLKEALIDAILQAIPGLGDRK
RIEVLSIYEGSINAVLRIHPPVADGPASSEGSDSPRQLTAAEALLALQEQLEEPLSVLRT
GPFGEYAAYATIGPGEGQQLLLRGIGSGPDVVPNLLLMCFKAASLGAPAAAASTE
- Download Fasta
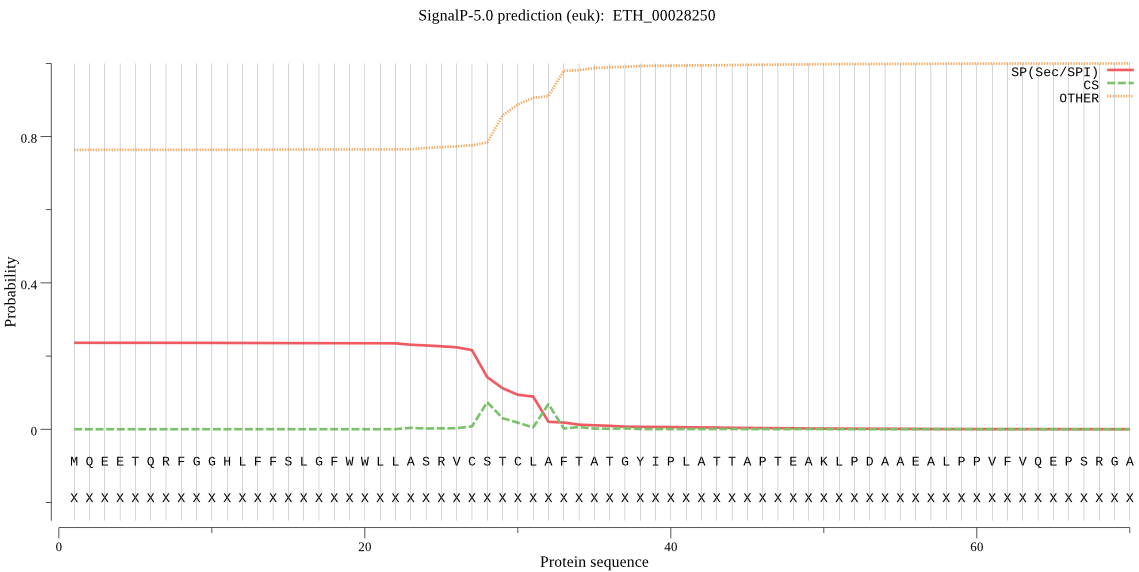
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00028250.fa
Sequence name : ETH_00028250
Sequence length : 1975
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.578
CoefTot : -0.155
ChDiff : -38
ZoneTo : 2
KR : 0
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.988 1.788 0.445 0.713
MesoH : 0.300 0.608 -0.111 0.328
MuHd_075 : 18.089 9.124 5.111 4.493
MuHd_095 : 29.608 16.164 7.811 5.437
MuHd_100 : 25.551 13.746 6.540 4.030
MuHd_105 : 22.602 10.387 4.999 3.090
Hmax_075 : 12.717 8.517 0.794 5.297
Hmax_095 : 17.150 11.900 2.164 5.381
Hmax_100 : 15.500 10.400 1.767 4.290
Hmax_105 : 8.600 5.000 -0.183 2.800
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.8979 0.1021
DFMC : 0.9142 0.0858
-
Fasta :-
>ETH_00028250
ATGCAAGAAGAGACGCAGAGATTTGGAGGCCACTTGTTTTTCAGCTTGGGGTTTTGGTGG
TTGCTTGCCAGCCGTGTTTGCTCAACATGTCTGGCCTTCACCGCAACAGGATATATACCC
CTAGCAACAACAGCGCCAACTGAGGCAAAGTTGCCAGACGCAGCCGAAGCACTTCCACCG
GTTTTCGTCCAAGAGCCTAGTCGTGGAGCCTTACCTGTGTGTGACTGCACTGGACTCCAT
CATGGAACTGTGCACGACGCCGCATGTTCCATAGGCTTTTCTGATGAAGCCACCGACTTT
TGCTTTATTCCGCCTTCCTACTGCGCAACTGTGGAATCCGGGGAGATGCTCACAGTGAAT
GGTAGTCGCTTGGAAGAATACTCTATACTCCTTGTTGACTGGGGGAAGACCAAGGAATCG
GCAGATTACTACGAGAAGGCCCTACCTAGGGCGTCGACGCCAACATGCCGCTGCTGGCCC
TTTAACGGTGTAATGGTGGCGCCGTGTGAGCAGCAGTGGCCTCCACAGGTTCGAGCCGAA
GGCACAGGCGGTAGCAACACAAGTGCAGGAGATTTATCAGCTGCCTCTGAATCAATCCTT
GCGGGTGGGGTACTTTCACGGCGCCAATCGGCATTGAAAAAATCATTTGTCTCTGGCGGT
GATGCTCCTGACGCAGCAATTCATCGAAATTTGTTCCACACGCCTATTCAGGCCACAGGC
CTCCAGCGGTTAAGCAAGTGGGCACGCCACTTGGCCGTTTGGAGACAGGCAATAACAGGA
GATAGCCATAGCTCAAAAGATGTAACTGCGCCTTCAGTGAATACCACGTGTCATAAGGGC
TGCGCAAGCTGCTTACAGCCTGGTCGAGATCGCTGCCTCACATGCTTCAATCAGTTGCCA
TATCCCTACCGGCCGTTACCGGCGGGTTCACACCCTCCTAACCCCAATAAGCTTCCCTCT
GACATTTACGGCTCTAATGCTTACAAAAAGCAGTATCATGCTCACATTGCCGCTCACCGA
GATGGTAGTGGTTACTGCATCCCATTGGCTTTCGGGGGCGATGCCCTTGCAGCGGCACAT
TCTTCATGTCTAGCTTCGGATTCATCATGCGTTAAGAAAAATGTGAGACCACTGGAGCAG
CCAGAGGGCGGGTTCTTGGAACTCAATAACGGCAGTCACTGCCACGCGTCCTGCGCTGCT
TGCCGTGCTGATTGCTGCCGTGGAAAGGCTGACAACTGTCTGGCGTGTACCGGGCCAAGC
CGGATATTCCATTCACTCTATGGAGATGGTACAGGGCGCTGTATAGATTTAAGGCTCCCC
GATGACCAAGTACAGACGACAAATCTTGTGGAAAAAGGACAAGATGAGGGAATGATGTAC
ATGCAGCAATTAGCGAACTTCGAGGGTGAAGAAACAGGCAGCTTTTCAGCCGTCCTTCAG
TCCATAATCAGTATCGCCTTTTCTAGCCCCGCTGCACCTGACACAGGGAAGCCCAAAGTC
GCAGCATCGTTGGATAAGAGCAGGAATAGAAAAGGCCCATCCTTTCTTCCACGACGATCT
ATTTGCGACTCATTACCTCAGCGCATTGGAGATATGCCGACTCGCGCCTGGTCTCAGCTC
ATGTTTGTTCATGCAGACAACAACTTGGAAGAATCTGCTCTATTAGACTTGGAAGAAATG
CTACATCCGTGGAGAGGCGAAGTAGTGAAACGATATGCTCGGGCCGCTATGCGAGGGCGC
GGCACGCCGCCTTTGCCCGTTGGTGGATCGGCACTGAGTGGTTCCACAGCCCAAGAGGCA
CTAGAGGACCTGTATCTTGTCGTTCTGATTGATCGTAGCAAGCAGACCACTTCAACAGAC
ATGGGGACAGTTCATGCATGTCCTGAGCTGGAATACTCCAGTTTAGGCCAAGGTGTCAGG
CAACTACCAGGAACAAGTGAAGTCGAGCTCAATGCCCAAATGGCATTTGAGCTACTGCGA
ATCCATTTGCAAGACGGCAGGCGCGAATGGCTGCTCCTTCGCAGTCTTGGGGAGGTTGAT
ATGAATGACCCTGGAGTTTTAAGAACTGCTGTGACGCGACTCCTCGACATATTTCCGTCG
AGGCACTACGCAGTCATCTTGTGGAATCACGGTTCGGCTTGGGCTGGGTTTGGTGATGAC
GAGAGCAACCCAAACAATCAGCCGATGAGCATTCATGATATCGCACTGGGACTCAAAAAG
TCCATGGATCCAACTTCTCTCACCCGGAAGCCACCTCCTCCCTACAGAACGGCTACAGCA
TTCGAGTATGGCGCACGCTTTGTTACCAGCTATGGCCTTCACCCGCCGTCCTCCAATGCC
TTGACGCTTGCTCTTATCGAAGTGAGAGCATTCAATGAGTTCAAGCAGAAGTTTGAAGAA
CTACTAGAGGGTCTTTATTCCTGCGGAGGAAGCAATATAACCCGACGAGTGAGGCGCGCG
CTTTCTCAAACTTTCAGCGTTAGAGGCTGCCGAATGCTGGGCTTATGCAGCTGTTATGAC
TTGAACGACTTTCTGGCAAATCTCTTGCAGCAGTTTGCAAGCGAGGAAAATGACTTAGGT
AGAATTAGCATGCAGTCACTTGGTCATGATGACGAGCTTTCGTGGGATGACAGTAATAGT
GAAATGCGAGACATAACGTACAGACGACTGCAGCATACAAATGGGCAACTGTACGCGATA
AATGCCTTGGGTGCTGCCCAAAGAGCTTTGACGGATAAAGTCGTCGCGGCTCGGGTGTTC
TTTAAGAGGATGGTCGTCGCTGAAGTTGGTAGCAGGGAACCAGGCAGATACGGGGGCCTG
AGCATTTACTTTCCTGATCCCAACATGGCAGTTAATTGCAAAACTCACCGAAGCGCAGCC
AGCTGGGCCCGGCGTTACACCAGCACCATTCGTACGAAGTTTTCCTCCTTCGTTTCATCC
GTGTTATCCAACCGAAAAGGTAGCGTGTGCTACGCTCCATGGGGAGGCACAGGAGATGCG
GCACAAGATGCTACGGGCACGATACCACAGCATGCCGCAGAAGCACCACAATGGTTTGGC
ATTCTGTGCAGTAGAGTCCTCAAATCGTACCTAGACAAGCATTTAGGGGCAGCTGTGGGG
ATCTCTGCTGTGCTTCCAGCGACGGTGGCTTCGGCTATGATGTTTAGGGGTTTCGCAGCT
AGTGTCGCGAATAACGGGAAGCCTGAAATAATTATCACAAGCACAGCCCAAGCGACAATG
ACGGATGCCGAAGTGAAAAGCTCAACAGTTCCCCTTACTTCCAGTGGGATGTCCGAAGAG
GCTCAAAGGTTGCATGCGCGATCTAAAGCTAATCTTCCGGGAAGCAGGGCGCACGACGCT
AGAGCTGAGCCATCAGACACTGCCGCTGCTCTCCGCGAATTTACAGTGGTGCAGGGGTGG
TGGGACACTCAAGTTTGGATCCTGAAACAGCAGCTGCGTCTCGACGATTCTCTGAAGGAC
CCAAGACCCCGGAGACAAACGCACAACGGCTCGGCCAAAGACGCGGACGTAGAAGCAATT
GTCGTCGCAATTCAAGATGGACCTGCATCATCCCGATTCGGAGCGGGGAGGGCAAGTCTT
TCATGCTGCCTGCTGACCGTTCTTCAGGGTACCACTTTCAGCTTTCCATTCCTCTTTTTT
AAAGATGCACTAGCTGCTGAGTGCTTGGAAGATTCTCTTCTGCATACGGAAGCTCTGAGT
GCGCCGAAAAGCCCTCAAAACTCCTTCGAATCAAGGTCAACGGCAGGGCAGGCAAAAGCG
CGCATGCTTCCAGCGGATCTTGCAGGTTTCAAAAAGCGCAGCCTGCAAGAACTTAGCAGC
TCTCCTGTCGAAGATATGAATAAGCAGGAAGCTTCCGTGAAGCGCCACATTTTGTCTTCT
GTGGTTCTTCCACTAGCTACGCCATCAGAGGACTTTCAAATAGTTCATGACGTAAGAGCT
AACTCGCAGGAGCAAGCATCAAGCAGCTCTTTGCAAGCCTCTTCATCGGATTTGCGCGGC
AGCCAAAAGCCCAAATTCGACGCCGAGTTGCAGGAGAAAGATGATGATGTCTCTAAAGTT
CCTCAGCCGCGTCAACTATCTAGCGTGCGAGCTGGGGCCGCGTCTTGGTTGCTTCACGAG
CGCCTAGGAAGGGAACCACAGAGGCACGTCGACAAAAAGCGGCAGTGTGGGGCAAGGGCT
TACCTGTTGGCAGAGTGGGATGAGAATCGGAGGGATTCCAGTGAACTGGTCCTCTACGTG
GTAGAGGACGAACAAGCAACGGAGTGGCCTCGCTCAAAAGGAGGGCTGCTGCGTCCCATC
GAGCACCGCCTCCGGAGGACAACAGTTTTTCAGCAATACTTCGAACAGGCTCCTTCGTCT
ATTCCGAATGCAAGAGGTGGAAGCCCCAGGAACAAAGGCACTTCACAGTCTGGGATAAGT
ACGAATGCTGCATATCAACCACGCAGTAGCGCAACAGGGCATCAGACGGGCTTTTCGTCA
GGCCATGGAGATGCTAGATTTTCGGAAAGCCTGTCGTTTACCAGAGCAGCCGGCAGTCAG
CATGAAGCCTTCTGGGGCCAACAAGCTGACCAAGGGTCCGAAGAAAACGTTGCGGCTGTT
GTGCTTGAGGAATCTCTTGGAGAGGCCACCTTCTTCTGGAACGTAGAGTCCGAAGTGGAC
AAACTTGTTCTCCAGCCACGTAGCATTTCCTCATTGCTTCGCGAGCGGCGCTCCAAAATG
AACGTTGAAGGAAGTCAAGTGCTGGAAAACACGGAAAGACCTGTTGTGGGTTTCATCGCA
CAAGATCTCTTTGACCAAAGGGCTGTGGCAGCAAGTGCAGTGCCGTTGACCGAACGGCAA
AGTCCTTCCTTTCCTCAGAGCCTCTGGGAATCCATAGAGCAAACGTTCGGCTTCGGAGAC
TATGCTCAAGAAACATTACAAGCCTCAAATGCCCTGCCATCTTCTTCCACGTGCCAGCCC
TACTGGCTAGGAGACGGCATCTGTGACGAGATATGCGACACAGCCGCGTATGAATTTGAT
GGAGGAGACTGCGTACACCTAAATGCTGCAGCGGAAGACCTCCTGGAGCAGCAGGAAGAA
ACGCCATTAGTGGCAACTATTTGGGAGGGTCCGAGAGGACGTTACGTCTGCAGGTGCCCA
AAGGGCTACACTGGAGATGGGACCTCTTGCGAAGATATCGACGAGTGTAAGGTCTTCGAG
GAAGCAGTCTGCCACGCCAAGGCCATATGCATCAACACCAAGGGCAGTTACAGGCCCAGC
AACCAGAGCTACAGCGGAAGTTGCATCGATATCGACGAGTGCTCTTCCGGACTCGCAAGC
TGCGCCTCTCCCTCCATCGCAACTTGTGTTAACACAAGTGGGGGATATTTCTGCAGTTGT
GTTGATGGGTATGAAGGTGATGGGCGCCAGTGCGTCGCCAGCCCCTCCACAGCTGAGGCA
GAAATTTCTATTGCTATGGACTACCATAAAACAGTTGCTGCTGCGGGAGGTATCCAACTG
CTTAAAGAAGCACTGATTGACGCAATCTTGCAGGCAATCCCAGGGCTGGGCGACAGGAAA
CGCATTGAAGTCTTGAGTATTTACGAAGGAAGCATCAATGCTGTTCTTCGGATTCATCCT
CCGGTGGCGGATGGACCAGCCTCCTCTGAAGGATCAGATAGCCCCCGGCAGCTCACAGCG
GCTGAAGCTCTGCTAGCTCTCCAGGAGCAGCTCGAGGAACCACTTTCAGTCCTCAGGACA
GGACCTTTTGGGGAATACGCTGCATACGCTACAATAGGTCCAGGAGAGGGTCAGCAACTC
CTGCTGCGCGGCATCGGGTCAGGCCCCGACGTGGTTCCTAATCTCCTCTTAATGTGTTTC
AAAGCCGCTTCTTTAGGAGCTCCTGCAGCTGCCGCTAGTACAGAGTAA
- Download Fasta
- title: Ca2+ binding site
- coordinates: D1770,E1773,N1791
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00028250 | 520 S | LPRRSICDS | 0.997 | unsp | ETH_00028250 | 520 S | LPRRSICDS | 0.997 | unsp | ETH_00028250 | 520 S | LPRRSICDS | 0.997 | unsp | ETH_00028250 | 968 S | RRYTSTIRT | 0.997 | unsp | ETH_00028250 | 1098 S | SSGMSEEAQ | 0.997 | unsp | ETH_00028250 | 1157 S | RLDDSLKDP | 0.997 | unsp | ETH_00028250 | 1171 S | THNGSAKDA | 0.996 | unsp | ETH_00028250 | 1322 S | VRANSQEQA | 0.995 | unsp | ETH_00028250 | 1334 S | SLQASSSDL | 0.995 | unsp | ETH_00028250 | 1341 S | DLRGSQKPK | 0.996 | unsp | ETH_00028250 | 1413 S | NRRDSSELV | 0.996 | unsp | ETH_00028250 | 1519 S | RAAGSQHEA | 0.995 | unsp | ETH_00028250 | 1729 S | GDGTSCEDI | 0.997 | unsp | ETH_00028250 | 1866 S | IEVLSIYEG | 0.992 | unsp | ETH_00028250 | 1888 S | DGPASSEGS | 0.994 | unsp | ETH_00028250 | 1894 S | EGSDSPRQL | 0.995 | unsp | ETH_00028250 | 218 S | KSFVSGGDA | 0.996 | unsp | ETH_00028250 | 264 S | GDSHSSKDV | 0.998 | unsp |


