

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
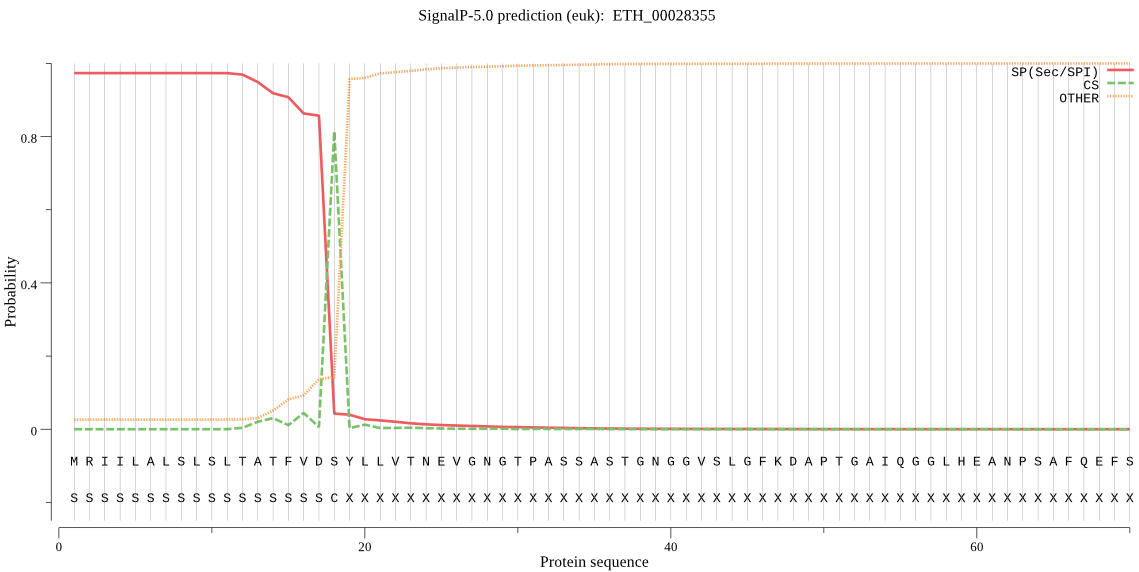
| ETH_00028355 | SP | 0.008852 | 0.990799 | 0.000349 | CS pos: 18-19. VDS-YL. Pr: 0.8847 |
MRIILALSLSLTATFVDSYLLVTNEVGNGTPASSASTGNGGVSLGFKDAPTGAIQGGLHE ANPSAFQEFSLIEEGAPIESKTGSVNEAERQKSTPEGGYLSAKRKSTQAKSSVNLTEVEE FASVVKIFVDAVKADFVSPWQMMAPKEQTGSGFVVEGRMIMTNAHLIADQTRVLVRRHGN PKRFLARVLAVCHECDLALVTVDDDVFWERIKPLAFGGVPQLRETVVVLGYPTGGDQLSI TEGVVSRVGVSMYAHSSLGLLTVQIDAPINPGNSGGPALAAGKVVGVAFQGFSEMQNVGY IVPFPVIRHFLNDIALHKQHTGIVSLGIKAQPMENEALKRFKGMDTLPPEALPENVTASG VLVVSVDKVRRTIYTQGKIALPLSRYSQIPESSSAGKRCVRYKAHVGPSIAAFDSSMAKC ELMKPILLVDQPVPRENDTTEHQRNVKEIAARVIWQPENWKPSTAASCSYCHHQQLIPRE KRNVLQNQKKVSQSGTPEAADVQRETGPMQWVKTIVQVEDVKGVADAEGSRLLSTVETKE KHLRRVSAQGSTSNREVNEESAPQSGQDARMASPLLRADIESAPQSGQDAKTASPQLGAD TAGVHESTSARSEPATLDLGVRKKAESAGALKYTPTNSSGHGSGDEDDERQKGQGRPDLR TQTGQTPVPKKRRSHTSNIRASLESLLREILRGSSKLRQVRNILLGRGGGDESDNLRGTF SATNTKRKAEQASNSNDNTQNEDALEKRPVVSGLKLAGDNTAGSAESNDSSPRSSNTSSK ELRSEERTATENDAEEKRNLEEKSQKYESHDNERETEDTALAVAVHPLVDNPYFNPAFLD EKEFLGFQEGDVILSIGNYSMDLERVSFHYVITQYFNGETTWAYVLRDNRVIKITVPLMT PNFKVPPFTWDMKPSYFVYGGFVFTPLSKVLLATKLRKEAPHMFGFLVQRLDYQEEAGDE FVVLSLILASDISVGYEVPPCIVHSVNGHHVRNMQDLVRFIEAKDGDFVELQLEANEADK LHVAIDRKKAAAIQQKVLKDHNIVSDRSPDLIRKKINSL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00028355 | 565 S | SAPQSGQDA | 0.992 | unsp | ETH_00028355 | 565 S | SAPQSGQDA | 0.992 | unsp | ETH_00028355 | 565 S | SAPQSGQDA | 0.992 | unsp | ETH_00028355 | 586 S | SAPQSGQDA | 0.991 | unsp | ETH_00028355 | 627 S | KKAESAGAL | 0.991 | unsp | ETH_00028355 | 674 S | KKRRSHTSN | 0.997 | unsp | ETH_00028355 | 682 S | NIRASLESL | 0.994 | unsp | ETH_00028355 | 764 S | NTAGSAESN | 0.992 | unsp | ETH_00028355 | 775 S | SPRSSNTSS | 0.995 | unsp | ETH_00028355 | 778 S | SSNTSSKEL | 0.998 | unsp | ETH_00028355 | 784 S | KELRSEERT | 0.997 | unsp | ETH_00028355 | 809 S | QKYESHDNE | 0.993 | unsp | ETH_00028355 | 93 S | ERQKSTPEG | 0.997 | unsp | ETH_00028355 | 534 S | SRLLSTVET | 0.997 | unsp |