-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00028760
MSSGTPVHHRHATPKPTVAERLHGHKEAPPQVIPMTNVMPGDSLFVTGDQFQSAMSVIRT
VIGGTGKSGATFPILPEDGSTMQAFLLRIERRYIQMGLKPR
- Download Fasta
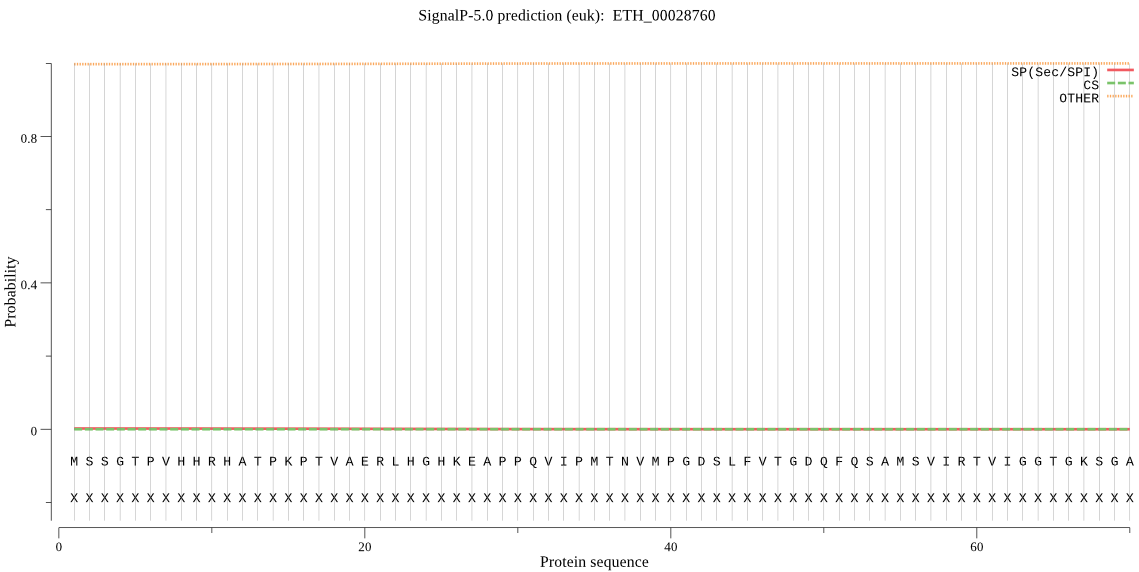
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00028760.fa
Sequence name : ETH_00028760
Sequence length : 101
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.271
CoefTot : -1.198
ChDiff : 4
ZoneTo : 19
KR : 2
DE : 0
CleavSite : 12
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 0.935 0.841 0.021 0.442
MesoH : -0.308 0.143 -0.352 0.196
MuHd_075 : 25.280 12.548 6.107 4.905
MuHd_095 : 11.356 5.992 1.698 2.108
MuHd_100 : 5.703 6.479 1.854 2.080
MuHd_105 : 2.535 7.082 2.431 2.265
Hmax_075 : 6.800 4.800 -0.765 2.140
Hmax_095 : 5.250 0.263 -2.214 1.173
Hmax_100 : 4.800 -0.400 -2.411 1.020
Hmax_105 : 7.600 4.400 -0.985 2.990
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9155 0.0845
DFMC : 0.8209 0.1791
-
Fasta :-
>ETH_00028760
ATGTCGAGCGGGACGCCGGTCCATCATCGCCATGCGACTCCGAAGCCCACCGTGGCAGAG
CGGCTGCATGGGCATAAGGAGGCGCCCCCGCAGGTGATACCTATGACCAATGTTATGCCA
GGGGACAGTCTCTTCGTCACGGGGGACCAATTCCAGTCCGCTATGAGTGTGATAAGAACT
GTTATAGGCGGGACCGGGAAATCAGGCGCAACGTTTCCCATACTACCAGAAGACGGCAGC
ACTATGCAGGCCTTTTTGCTGCGCATCGAGCGCCGGTACATCCAGATGGGATTGAAGCCG
CGCTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00028760 | 13 T | HRHATPKPT | 0.996 | unsp | ETH_00028760 | 56 S | QSAMSVIRT | 0.996 | unsp |


