-
Computed_GO_Component_IDs: GO:0016021
-
-
Computed_GO_Function_IDs: GO:0004190
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
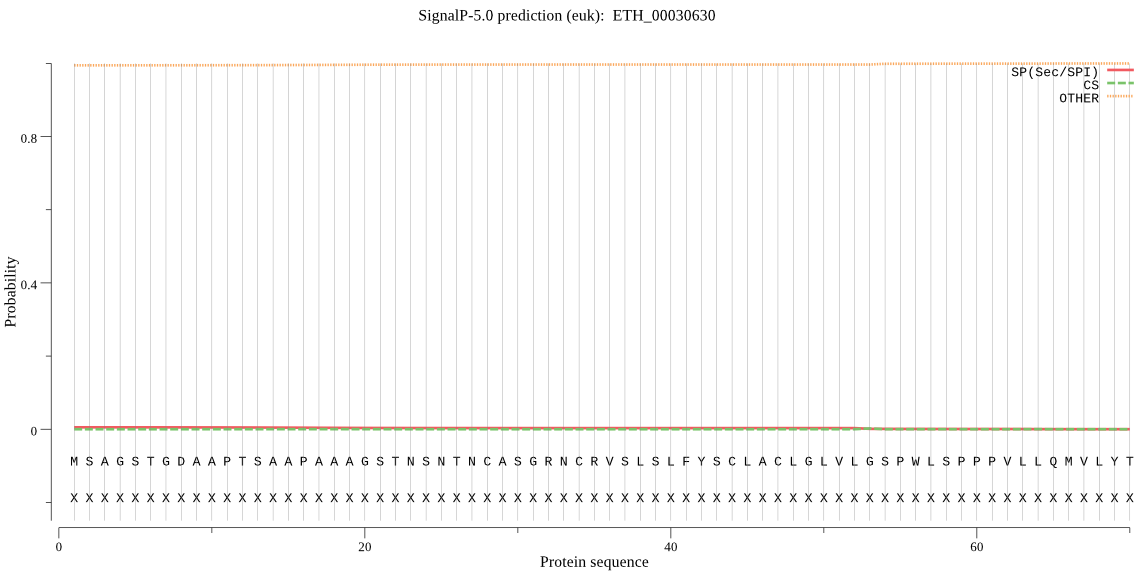
ETH_00030630 | OTHER | 0.999912 | 0.000087 | 0.000001 | |
No Results
-
Fasta :-
>ETH_00030630
MSAGSTGDAAPTSAAPAAAGSTNSNTNCASGRNCRVSLSLFYSCLACLGLVLGSPWLSPP
PVLLQMVLYTAPILYIGSHLSLKQNEVDAITGERLNKGEAMDRTDAMLFPVFGSIALFSL
YLAYKFLGAGWVNMLLTFYLTGVGLLALGGTIFSVSRPLCPAWLYDDSWLVLRPRGPFLW
VRKWLSGPTPSEATRMEHEEHQEEQQEKQQKKQQGQETKLAVDWSWRFSPLWLASHLLAL
AICILWLITKHWALHNILAIAFCTQAIAIVSVGSFGVASILLCGLFVYDVFWVFGTEVMV
SVAKAFEGPAKLMFPVQLSPLQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKAL
AVDIHQRFPKFYFCVVLAFYELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNG
KVKEVLAYKEEDEEGSSILRASALETTEKKEN
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00030630.fa
Sequence name : ETH_00030630
Sequence length : 452
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.733
CoefTot : -0.699
ChDiff : -2
ZoneTo : 85
KR : 3
DE : 1
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.112 2.318 0.532 0.823
MesoH : 1.369 1.650 0.141 0.636
MuHd_075 : 17.872 22.779 7.166 4.737
MuHd_095 : 22.842 15.843 5.897 6.200
MuHd_100 : 26.825 16.564 7.395 6.333
MuHd_105 : 27.619 20.900 9.418 5.551
Hmax_075 : 12.250 19.600 2.423 5.973
Hmax_095 : 12.075 21.263 5.954 5.050
Hmax_100 : 16.700 18.400 5.789 5.530
Hmax_105 : 15.900 19.367 6.972 4.707
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.5149 0.4851
DFMC : 0.2973 0.7027
This protein is probably imported in chloroplast.
f(Ser) = 0.1529 f(Arg) = 0.0235 CMi = 1.48571
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>ETH_00030630
ATGTCAGCAGGTTCAACAGGAGACGCGGCTCCTACGTCTGCTGCTCCTGCAGCAGCAGGC
AGTACCAACAGTAACACCAACTGCGCCAGTGGCAGGAACTGCCGTGTGTCACTGAGCTTG
TTTTACAGCTGCTTAGCCTGCTTGGGTCTTGTTTTGGGATCCCCCTGGCTGAGTCCCCCG
CCTGTGCTGCTCCAGATGGTGCTTTATACGGCCCCCATACTTTATATAGGGAGCCACTTG
TCTTTGAAGCAAAACGAAGTAGACGCAATAACGGGCGAGAGGTTAAACAAAGGGGAGGCC
ATGGACAGAACTGACGCCATGCTTTTTCCTGTTTTCGGCTCCATAGCACTCTTCAGCTTG
TACCTTGCCTACAAGTTCTTGGGAGCTGGGTGGGTGAATATGCTCCTGACGTTCTACTTG
ACGGGCGTAGGTCTTCTTGCGTTAGGGGGAACAATCTTCTCTGTGTCCAGGCCTCTCTGT
CCAGCCTGGCTATATGATGACAGTTGGCTTGTCCTCCGTCCCAGAGGTCCTTTCCTTTGG
GTCCGAAAGTGGCTCAGCGGCCCAACGCCCTCAGAAGCGACACGCATGGAACACGAAGAA
CACCAAGAAGAGCAACAGGAGAAACAGCAGAAGAAGCAGCAGGGACAGGAAACCAAGCTA
GCCGTGGACTGGAGTTGGCGATTCAGTCCTCTTTGGCTCGCTTCGCATCTTCTAGCTTTG
GCCATATGCATTCTGTGGCTTATAACGAAGCACTGGGCGCTACACAACATTTTGGCGATT
GCATTTTGCACTCAGGCGATAGCAATAGTGAGCGTGGGAAGTTTCGGAGTTGCTAGCATT
CTACTGTGCGGCCTCTTCGTATATGATGTGTTCTGGGTGTTTGGGACTGAAGTGATGGTT
TCAGTGGCAAAGGCTTTTGAGGGTCCAGCCAAACTGATGTTTCCAGTGCAGCTGTCGCCA
TTGCAGTACAGCATTCTCGGCTTGGGTGACGTGGTTATTCCTGGTGTCTTGATAGCCATG
TGCCTACGCTTCGATCTGTTTCTCTACCAAAAGCAGCAAAACGCAGCAACGAAGGCTCTG
GCGGTGGACATACACCAGCGTTTCCCTAAGTTCTACTTTTGCGTTGTACTTGCGTTCTAT
GAGTTGGGCCTGCTGACGACGGGCCTCGTAATGCTCTACGCGCAGCATCCGCAGCCTGCG
CTTCTTTATCTTGTCCCTTACTGTCTCTTTAGTCTCTTTGGCGCCGCTGCTCTCAACGGA
AAAGTTAAAGAAGTGCTGGCGTACAAGGAGGAAGACGAGGAGGGGAGCAGCATCCTGCGA
GCGTCTGCGCTGGAGACAACAGAGAAGAAAGAGAACTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00030630 | 5 S | MSAGSTGDA | 0.991 | unsp | ETH_00030630 | 442 S | ILRASALET | 0.993 | unsp |


