-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00031605
MRLHNGRVIVPTPQETSIISANKMPSGTPAHHRHATPKPTVAERPHGHREAPQQMISMTH
VMPGDSPFVTLDQFQSAMSVIRTAIGGTVESGATIPILPEDGSTMQAFLLRVERRYTQMG
LEPREWGNALIDHLVGPALTYWMYLRRTIDLCDWATVRRRLLERFDKTMSQSQLLTEFAR
VRWNGNPNEYLDRFAAVAERGLGVAPDELADYYCTRLPTDLHLPITNNGQVKYQSWEQAA
TAAARLYEPTQSVLELRQRTSRAIRAAIQANGPRRRQEIENRPGGTHANCSECQGRGHPT
RLCPSKGERTKRPGEICKKCGGVGHYARDCPTYERGRPEPGDKSSREPTRPSTEKTERLN
RGPRRGWRRAF
- Download Fasta
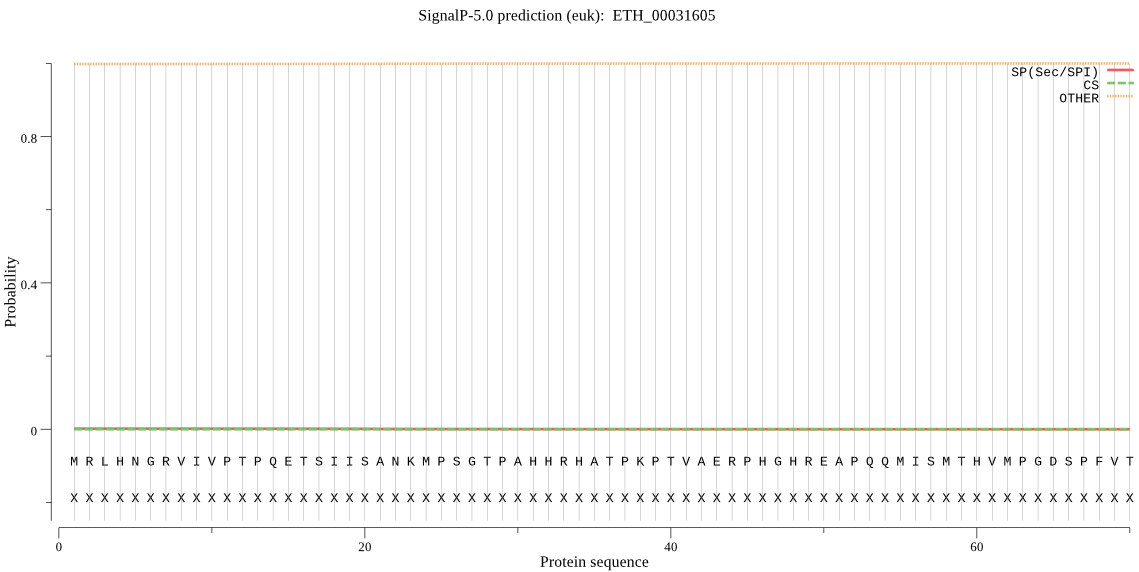
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00031605.fa
Sequence name : ETH_00031605
Sequence length : 371
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.942
CoefTot : -2.429
ChDiff : 16
ZoneTo : 42
KR : 5
DE : 1
CleavSite : 35
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.000 0.971 0.092 0.551
MesoH : -0.375 0.159 -0.363 0.186
MuHd_075 : 29.971 17.513 8.535 6.966
MuHd_095 : 32.922 12.218 8.079 4.639
MuHd_100 : 22.234 10.805 5.390 4.468
MuHd_105 : 21.218 10.998 6.522 4.815
Hmax_075 : 3.733 11.550 -2.284 4.611
Hmax_095 : 11.700 9.200 1.406 3.610
Hmax_100 : 6.400 0.400 1.430 1.730
Hmax_105 : 7.700 9.300 2.072 1.808
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.2957 0.7043
DFMC : 0.4338 0.5662
This protein is probably imported in mitochondria.
f(Ser) = 0.0714 f(Arg) = 0.0714 CMi = 0.42017
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>ETH_00031605
ATGCGTCTCCACAACGGACGCGTTATCGTTCCCACACCGCAGGAGACTTCAATAATATCA
GCAAACAAAATGCCGAGCGGGACACCGGCCCATCATCGCCATGCGACTCCGAAGCCCACC
GTGGCAGAGCGGCCGCATGGGCATAGGGAGGCGCCCCAGCAGATGATATCTATGACCCAT
GTTATGCCAGGGGACAGTCCCTTTGTCACGTTGGACCAATTCCAGTCCGCTATGAGTGTG
ATAAGAACTGCTATAGGGGGTACCGTGGAATCGGGCGCAACGATTCCCATACTACCAGAA
GACGGCAGCACTATGCAGGCCTTTCTGCTGCGCGTCGAGCGCCGGTACACCCAGATGGGA
TTGGAGCCGCGCGAGTGGGGAAATGCACTTATTGACCACCTTGTGGGCCCGGCTCTAACA
TATTGGATGTATCTACGGAGAACTATTGACCTATGCGACTGGGCGACAGTACGGCGCAGG
CTTTTGGAGCGGTTTGACAAAACAATGTCGCAGAGTCAATTGTTAACGGAGTTTGCTAGA
GTACGGTGGAATGGTAACCCGAATGAATACCTGGACCGGTTTGCGGCTGTAGCGGAGCGT
GGTTTGGGTGTTGCCCCCGACGAACTCGCTGACTATTACTGCACCAGGCTACCGACAGAC
CTCCATCTCCCAATAACCAATAACGGACAGGTGAAATATCAGTCGTGGGAACAAGCGGCG
ACAGCTGCAGCACGGCTCTACGAGCCCACGCAGAGTGTGCTGGAACTCCGACAAAGGACC
AGTCGAGCCATTAGGGCTGCTATCCAAGCTAATGGACCCCGTAGGCGCCAGGAAATTGAA
AATCGACCTGGGGGAACCCACGCGAACTGCTCTGAGTGTCAGGGTCGCGGACATCCGACA
CGTCTCTGCCCTAGCAAAGGGGAACGTACCAAACGTCCGGGGGAAATATGCAAGAAGTGT
GGCGGTGTGGGACACTATGCTCGGGACTGTCCTACGTATGAACGAGGCCGGCCCGAGCCG
GGGGACAAGAGCAGTAGGGAGCCCACCCGTCCTAGCACTGAGAAGACGGAACGATTAAAC
AGGGGGCCTAGGCGTGGATGGAGACGAGCATTCTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00031605 | 344 S | PGDKSSREP | 0.994 | unsp | ETH_00031605 | 344 S | PGDKSSREP | 0.994 | unsp | ETH_00031605 | 344 S | PGDKSSREP | 0.994 | unsp | ETH_00031605 | 345 S | GDKSSREPT | 0.992 | unsp | ETH_00031605 | 352 S | PTRPSTEKT | 0.997 | unsp | ETH_00031605 | 36 T | HRHATPKPT | 0.996 | unsp | ETH_00031605 | 79 S | QSAMSVIRT | 0.995 | unsp |


