

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
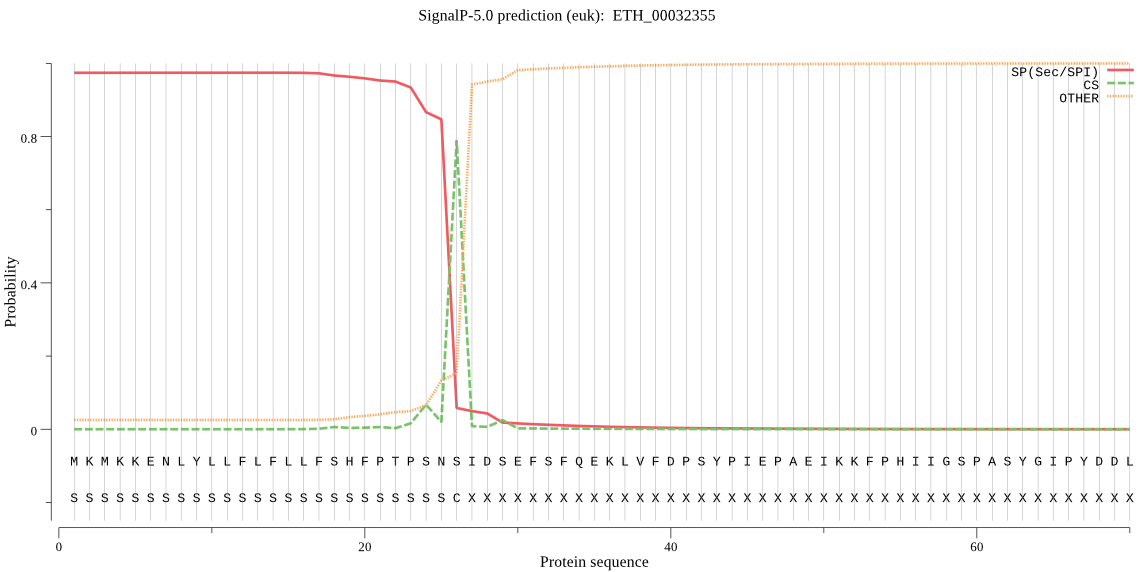
| ETH_00032355 | SP | 0.067866 | 0.932132 | 0.000002 | CS pos: 26-27. SNS-ID. Pr: 0.9194 |
MKMKKENLYLLFLFLLFSHFPTPSNSIDSEFSFQEKLVFDPSYPIEPAEIKKFPHIIGSP ASYGIPYDDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQG MNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSSNSNSNANNNNANA ISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLRE AAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEKR KRMWIGDE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00032355 | 194 S | SSSSSSSSS | 0.994 | unsp | ETH_00032355 | 194 S | SSSSSSSSS | 0.994 | unsp | ETH_00032355 | 194 S | SSSSSSSSS | 0.994 | unsp | ETH_00032355 | 197 S | SSSSSSKSN | 0.997 | unsp | ETH_00032355 | 198 S | SSSSSKSNF | 0.991 | unsp | ETH_00032355 | 293 S | KPRHSSRLY | 0.997 | unsp | ETH_00032355 | 137 T | RSEGTPSEA | 0.992 | unsp | ETH_00032355 | 193 S | SSSSSSSSS | 0.991 | unsp |