

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00040925 | OTHER | 0.999965 | 0.000034 | 0.000001 |
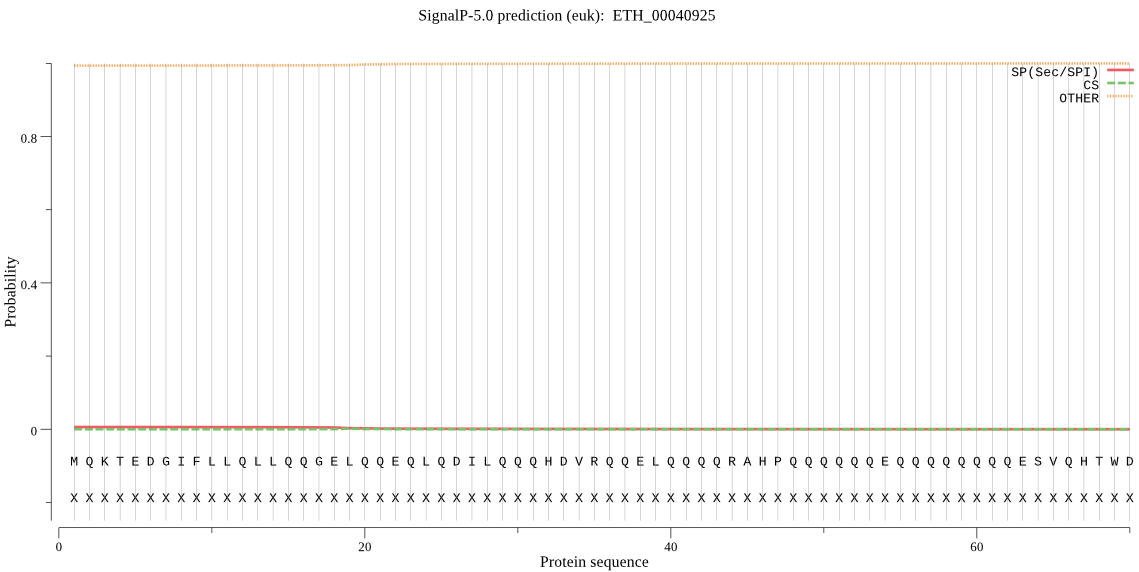
MQKTEDGIFLLQLLQQGELQQEQLQDILQQQHDVRQQELQQQQRAHPQQQQQQEQQQQQQ QQESVQHTWDAAARAALLMEGKHYEQFLLLSPSQLTATGLKELCLGYTPAAAAATAAAAA TAAPTVALPLYVPCVLFRNNHFSVIVRQPLPSCQQKLLQQLEQQQQQEDVLQEQQQQQQQ ECAIYCLCTDAAFSDEPQIVWEILIDVKGDNLFCNDKFLPLQYTQEGESSSCSSAASLGG ALQQEQ
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| ETH_00040925 | 194 S | DAAFSDEPQ | 0.992 | unsp |