-
Computed_GO_Component_IDs: GO:0005737
-
Computed_GO_Components: cytoplasm
-
Computed_GO_Function_IDs: GO:0008239
-
-
Computed_GO_Process_IDs: GO:0006508
-
Computed_GO_Processes: proteolysis
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
|
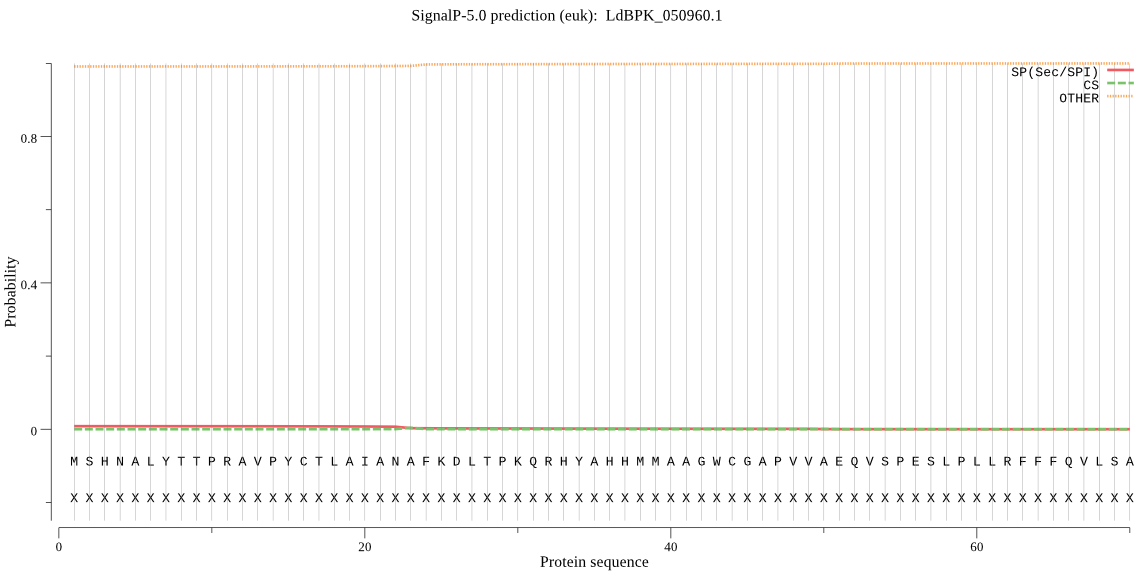
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
LdBPK_050960.1 | OTHER | 0.944542 | 0.016630 | 0.038828 | |
No Results
-
Fasta :-
>LdBPK_050960.1
MSHNALYTTPRAVPYCTLAIANAFKDLTPKQRHYAHHMMAAGWCGAPVVAEQVSPESLPL
LRFFFQVLSAQPLDTFKANSVNAGVDPDEVNQFLEYFAMVYSNMGNYLSFGDTKFVPSIP
KERFAKIVASADGSPAVDAQLLDAIYNLDDDKLTLDFPPKGLTRYYSPNVTREDAAVAND
FLASKKMDGVNTRVFKEEDGTLVIRVAAATERTVPAEKFNGRTIAIYYGDYKEEMARVVA
ELRKAQPYAENETEVRMLNHYIEHFQYGDVDVHKESQKEWVKDVGPTVETNIGFIESYRD
PSGVRAEWEGFVAVVNKEQSKMYGALVAQGEKFVAELPWGKAFEKDVFSSPDFTSLDVLG
FASSGIPAGINIPNYDDIRQTVGFKNVYLSNVVSAITFKDKLNYITEADWELYKASILTA
TSVNVGIHELLGHGTGKLLSENSDGTFNFDEKTVDPISGKPVATWYKPGDTYSSVFGGLG
SSYEECRAEAVSLYLCLLPDLLEIFNLQTTKEQQDVIYVCWLNMVRAGLVGLEFYTPEKQ
QWRQAHMRARFCILQALARAPNPIVQITENAKEGVLITLDRERIATDGRQAIGDLLVNLN
VNKATADAKRGSAYFENMTVVSDQYVHYRDIIMARRKPRKQYVQPHTFISGDTVEVREFA
GSVEGVVESFVTRHREIPL
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/LdBPK_050960.1.fa
Sequence name : LdBPK_050960
Sequence length : 679
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.150
CoefTot : -1.668
ChDiff : -19
ZoneTo : 50
KR : 4
DE : 1
CleavSite : 35
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.118 1.641 0.133 0.557
MesoH : -0.310 0.314 -0.322 0.251
MuHd_075 : 30.742 20.205 9.381 6.084
MuHd_095 : 37.614 25.982 11.442 7.194
MuHd_100 : 40.400 27.007 11.995 8.347
MuHd_105 : 31.000 22.216 9.756 6.893
Hmax_075 : 13.300 15.100 3.917 5.553
Hmax_095 : 16.275 21.400 5.082 5.495
Hmax_100 : 17.400 21.400 5.082 6.330
Hmax_105 : 14.700 19.133 5.023 3.680
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.7855 0.2145
DFMC : 0.7952 0.2048
-
Fasta :-
>LdBPK_050960.1
ATGTCGCACAACGCGCTTTACACTACCCCTCGTGCGGTACCGTACTGCACCCTGGCCATC
GCCAACGCCTTCAAGGACCTGACCCCGAAGCAGCGTCACTACGCCCATCACATGATGGCG
GCGGGGTGGTGCGGCGCCCCGGTGGTGGCTGAGCAGGTGAGTCCCGAGTCTCTGCCGCTC
CTGCGTTTCTTCTTCCAGGTGCTCAGTGCGCAGCCGCTGGACACCTTCAAGGCGAACAGC
GTCAACGCCGGCGTCGACCCCGATGAGGTGAACCAGTTCCTGGAATACTTCGCGATGGTC
TACTCGAACATGGGCAACTACCTCTCCTTCGGCGACACCAAGTTCGTCCCATCGATTCCG
AAGGAACGCTTTGCGAAGATCGTGGCGAGCGCGGACGGGTCTCCGGCGGTGGATGCGCAG
CTGCTGGACGCCATCTACAACCTGGACGACGACAAGCTGACGCTGGACTTCCCTCCCAAA
GGGCTTACGCGGTACTACAGCCCAAACGTCACCCGCGAAGACGCCGCTGTCGCGAACGAC
TTCCTGGCGTCGAAAAAGATGGACGGCGTCAACACACGCGTGTTCAAGGAGGAAGACGGC
ACCCTCGTCATCCGCGTCGCGGCGGCGACCGAGAGGACGGTGCCGGCGGAGAAGTTCAAC
GGACGCACCATTGCAATTTACTATGGCGACTACAAGGAGGAGATGGCCCGCGTCGTCGCG
GAGCTGCGCAAGGCGCAGCCGTACGCGGAGAACGAGACGGAGGTGCGTATGTTGAACCAC
TACATCGAGCACTTCCAGTATGGCGACGTGGACGTGCACAAGGAGAGCCAAAAGGAGTGG
GTGAAGGATGTGGGGCCGACGGTGGAGACGAACATCGGCTTCATCGAGTCGTACCGCGAC
CCCTCCGGCGTGCGTGCGGAGTGGGAAGGCTTCGTGGCGGTGGTGAACAAGGAGCAGTCG
AAGATGTACGGCGCACTGGTGGCACAGGGCGAGAAGTTCGTTGCTGAGCTGCCGTGGGGC
AAGGCGTTCGAGAAGGACGTCTTCTCCAGCCCGGACTTCACCAGCCTTGACGTCCTGGGC
TTCGCGAGCAGTGGCATTCCGGCTGGCATCAACATCCCCAACTACGACGACATTCGCCAG
ACCGTTGGCTTCAAGAACGTGTACCTGTCCAACGTGGTGAGCGCGATAACCTTCAAGGAC
AAGCTCAACTACATCACAGAGGCGGACTGGGAGCTGTACAAGGCGAGCATTCTCACCGCG
ACCTCGGTGAACGTCGGCATTCACGAGCTGCTGGGCCACGGCACAGGCAAGCTGCTGTCG
GAGAACAGCGACGGCACCTTCAATTTTGACGAGAAAACGGTGGACCCAATCAGCGGCAAG
CCCGTTGCCACGTGGTACAAGCCCGGCGACACGTATTCGAGCGTGTTTGGCGGTCTTGGC
AGCTCGTACGAGGAATGCCGCGCTGAGGCGGTATCGCTGTACCTCTGCTTGCTGCCAGAC
CTGCTGGAGATCTTCAACCTCCAGACAACGAAAGAGCAGCAGGACGTCATCTATGTCTGC
TGGCTGAACATGGTGCGCGCCGGCCTGGTCGGGCTCGAGTTCTACACCCCGGAGAAGCAG
CAGTGGCGCCAGGCTCACATGCGGGCACGTTTCTGCATTCTCCAGGCGCTGGCGCGTGCA
CCGAACCCGATTGTGCAGATCACCGAGAACGCCAAGGAGGGCGTCCTCATCACACTCGAC
CGCGAGCGCATCGCCACAGACGGGCGCCAGGCGATTGGGGACCTGCTCGTGAACCTCAAT
GTTAACAAGGCGACGGCCGACGCGAAGCGCGGCAGCGCCTACTTTGAGAACATGACGGTC
GTGAGCGACCAGTACGTGCACTACCGCGACATCATCATGGCGCGCCGCAAGCCGCGCAAG
CAGTACGTCCAGCCGCACACCTTCATCAGCGGGGATACGGTGGAGGTGCGGGAGTTTGCC
GGATCGGTAGAAGGGGTCGTGGAATCCTTCGTTACCCGGCACCGCGAGATCCCGCTGTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
LdBPK_050960.1 | 302 S | YRDPSGVRA | 0.996 | unsp | LdBPK_050960.1 | 302 S | YRDPSGVRA | 0.996 | unsp | LdBPK_050960.1 | 302 S | YRDPSGVRA | 0.996 | unsp | LdBPK_050960.1 | 276 S | VHKESQKEW | 0.997 | unsp | LdBPK_050960.1 | 297 S | GFIESYRDP | 0.992 | unsp |


