

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_070100.1 | OTHER | 0.999997 | 0.000003 | 0.000000 |
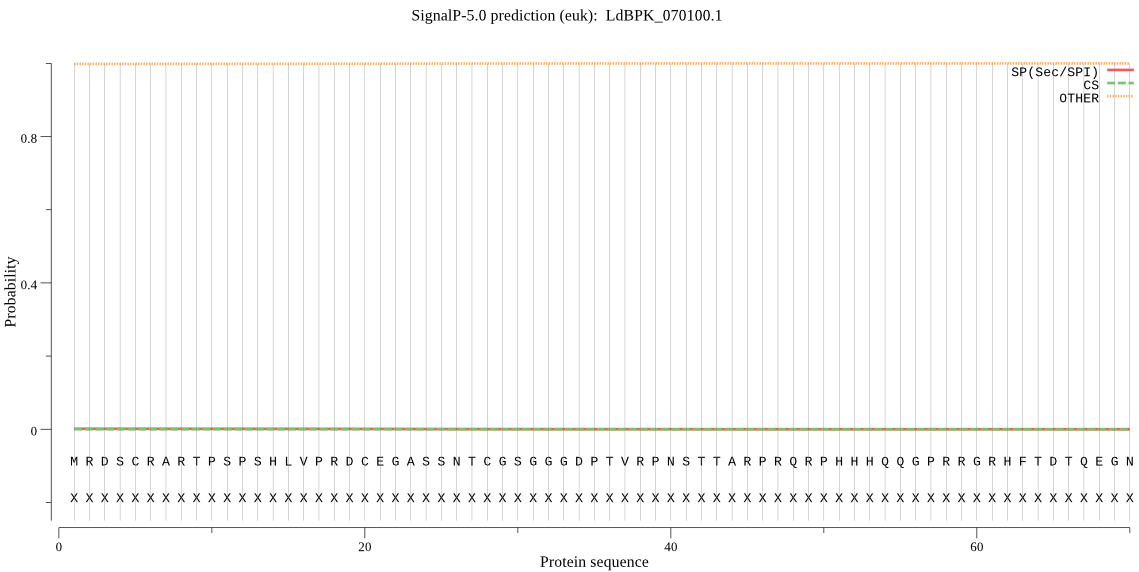
MRDSCRARTPSPSHLVPRDCEGASSNTCGSGGGDPTVRPNSTTARPRQRPHHHQQGPRRG RHFTDTQEGNGKSSRSSRGSVGGCVVPMWMNAPPNVVGGVERMANNSHQSSLSSFAAAVV NRPPGTSTSPTTQHKQWEAAARSTSGAASTSLAAAVAFGPSACVVASGAAVSGTQRSDHS PTGTAARVAQMPLRGAPSAAATPQHDCPSRDSSGYPQGSSPTSLRLHATASTATGTATCG NLADAQRRRRLKPPPIMSDVASASGRRSCSCPSTRSATAIEATIASSPLATYTVVAASPR AAAAVTHPARTLRSAGSPASPSDMLPQGPVPPPNPDFKPLGLWSTSPASAAASTSATTLE LKASTATASGADVGSLLTHEMDGGVVIVDPHQTPDDLVCGVCMCVCRQPTAAACGHLFCR RCLQSWMQENRTATCPLDRTPIRVELLHTDARAQRQINALRCRCPASLSRASQWALRQLG SASHVLGDRAVEDEADNDDARGDERARREHPRCTWAGCVSDAEAHLRQCPYIIIECPFAK RGCSAEMPRVDMAGHLKSGVADHLLLVSQALDASTEQYRVLQGEVEVLRQRCPMHYPVAL PDVPTGGPAGISEPAVSLYSLSLHGADVPAAALAPALVIAGGGGSSSGATPIAITAFPEE VSPESGSPTRVVGPFGHGASAAVDYPFPVFSHSQPPYPPPHHGSSTIATQTNVRRHAATD DMLLVAAHRMSSGHDATMPMPADTTALCTSPHFGAPPTSVSGANTAAAAPTFALPAGAAA PIPAQVASAPPRAAPVVGSGHCVDRFVWVITDMASRQAPCYSRSFTSHGLSWYVGIDTTA SWEQCGVYLFTEGHEHRVDFRVILYHEDPARDVVHVVRDWQEDYIGKGWGPLRFINRFTL EQDGFLVSGCLRVGIEVVSGPY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_070100.1 | 76 S | KSSRSSRGS | 0.995 | unsp | LdBPK_070100.1 | 76 S | KSSRSSRGS | 0.995 | unsp | LdBPK_070100.1 | 76 S | KSSRSSRGS | 0.995 | unsp | LdBPK_070100.1 | 77 S | SSRSSRGSV | 0.993 | unsp | LdBPK_070100.1 | 80 S | SSRGSVGGC | 0.994 | unsp | LdBPK_070100.1 | 180 S | RSDHSPTGT | 0.995 | unsp | LdBPK_070100.1 | 213 S | SRDSSGYPQ | 0.993 | unsp | LdBPK_070100.1 | 264 S | VASASGRRS | 0.99 | unsp | LdBPK_070100.1 | 298 S | VVAASPRAA | 0.996 | unsp | LdBPK_070100.1 | 320 S | GSPASPSDM | 0.996 | unsp | LdBPK_070100.1 | 393 T | DPHQTPDDL | 0.994 | unsp | LdBPK_070100.1 | 544 S | KRGCSAEMP | 0.992 | unsp | LdBPK_070100.1 | 662 S | PEEVSPESG | 0.993 | unsp | LdBPK_070100.1 | 665 S | VSPESGSPT | 0.992 | unsp | LdBPK_070100.1 | 732 S | HRMSSGHDA | 0.997 | unsp | LdBPK_070100.1 | 11 S | ARTPSPSHL | 0.993 | unsp | LdBPK_070100.1 | 41 S | VRPNSTTAR | 0.993 | unsp |