

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
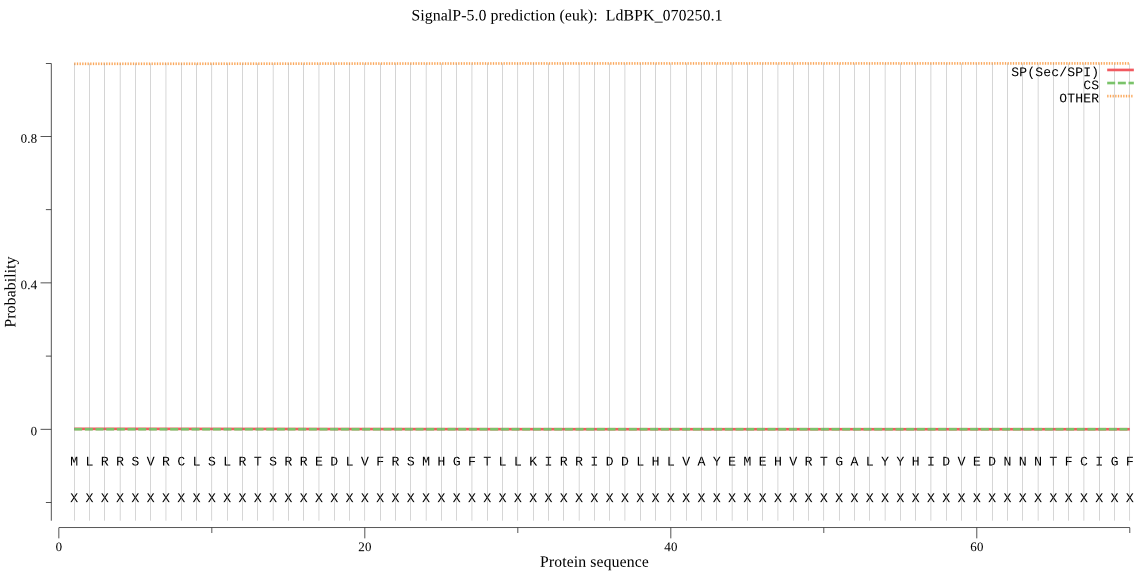
| LdBPK_070250.1 | OTHER | 0.685849 | 0.000073 | 0.314079 |
MLRRSVRCLSLRTSRREDLVFRSMHGFTLLKIRRIDDLHLVAYEMEHVRTGALYYHIDVE DNNNTFCIGFRTPAENNKGTSHVLEHTTLCGSKKYPVRDPFFMMLKRSLSSFMNAMTGSD YTLYPFSTTNRKDFQNLLDVYLDAVLHPLLREEDFKQEGHRVELEDKSGDSEDAAAQPAK RTRRLINNGVVFNEMRGVVSDPSNHFVHSLMRAMLPHTHYTYISGGYPPDILGLSYDELL SFQRRHYHPSNSITFTYGNLHPESHMEALDSYFADFERAAPVVVPTLADEHRFTEPQLVH LEGPLDAMGNPRRQKRVAVSYAVPKENNKLEDVVALSVLDSLLSSGPSSPMFKNLIESQI GSKYAPMQGYAFYLSSPIITYGVAGMDEGRADAEADVLQAVESALRTVQRDGFDERRVRS VIFQEELQHRHRSADYGLNTCTGLCAMGLCRAQNNPLDFINWLPHLRRLADDNAASLLPR IETHLLSNPHRAVVSVSAKKEYLNRLQDQLKEADEAVNASATEADKDRVEKETKEGLQRL RAPQPHDVLPTLRIEDIPTESLAEPVPCRSSLSSANGQVYTITHQTNGLVYVHGLIPFNT SLTSAMEHGELGQVPQSVMLLESLIGRTGAGKLSYKDHSIAVKLACSGFGFEPLLNESYS HKSTTITGTSYSFYTTKEKLKEALDLLSVTLLEPRFSADDADVYSRALSNLKMACSSVIQ SLQAEGNRYAVIRAVGELTRRGELREHWWGLSQSAHASEMLEKLQGSPEVSRETVSALLA NYAVFAQEMAADMSRSLVWATCEDAHREEVERMLKEFLDAFPRTDSAARTHLFLPPRSTE KGVQQIIKKLPIDTSFVGLAMPNKLKWESPDQARVRVGCTLLCNEYLHRRVREEGGAYGS NCTATLHGEVGGVSMSSYRDPSPELTAKAFLEAGDWLSDRKNVTAERVSEAKLRLFSSID SPYAADSYGEAYFYNDLRQDTKQALRDALLSVTAEDVVNVAHYFTPQSTTIISILRPAGE SSDPAAVSPEVS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_070250.1 | 634 S | AGKLSYKDH | 0.997 | unsp | LdBPK_070250.1 | 634 S | AGKLSYKDH | 0.997 | unsp | LdBPK_070250.1 | 634 S | AGKLSYKDH | 0.997 | unsp | LdBPK_070250.1 | 660 S | NESYSHKST | 0.991 | unsp | LdBPK_070250.1 | 697 S | EPRFSADDA | 0.997 | unsp | LdBPK_070250.1 | 767 S | KLQGSPEVS | 0.994 | unsp | LdBPK_070250.1 | 839 T | PPRSTEKGV | 0.993 | unsp | LdBPK_070250.1 | 917 S | VSMSSYRDP | 0.997 | unsp | LdBPK_070250.1 | 922 S | YRDPSPELT | 0.997 | unsp | LdBPK_070250.1 | 949 S | AERVSEAKL | 0.991 | unsp | LdBPK_070250.1 | 957 S | LRLFSSIDS | 0.997 | unsp | LdBPK_070250.1 | 961 S | SSIDSPYAA | 0.993 | unsp | LdBPK_070250.1 | 967 S | YAADSYGEA | 0.993 | unsp | LdBPK_070250.1 | 14 S | SLRTSRRED | 0.997 | unsp | LdBPK_070250.1 | 420 S | RRVRSVIFQ | 0.991 | unsp |