

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_110630.1 | OTHER | 0.999060 | 0.000479 | 0.000461 |
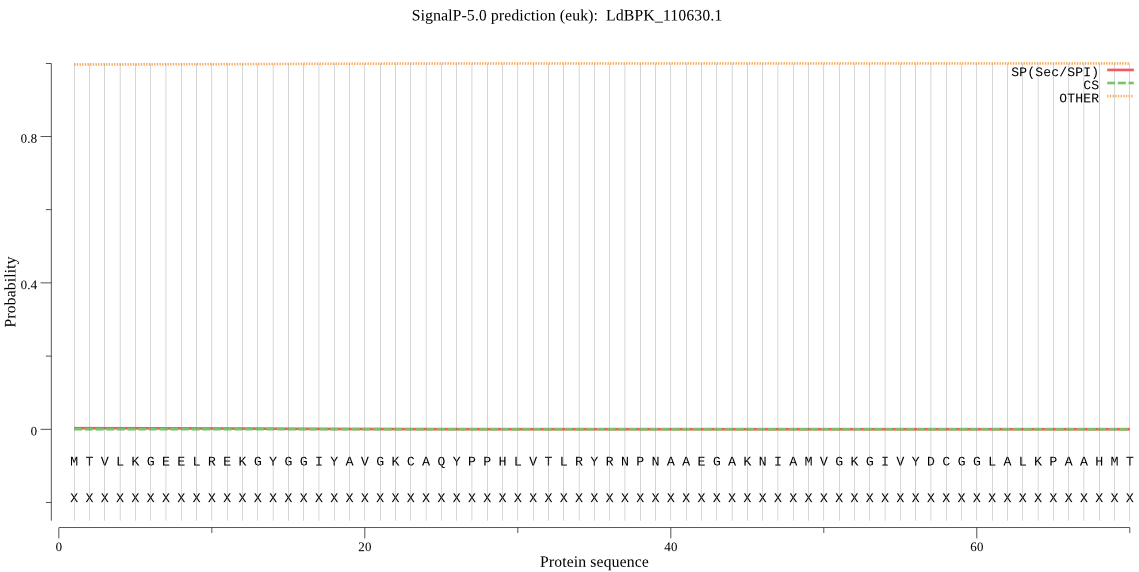
MTVLKGEELREKGYGGIYAVGKCAQYPPHLVTLRYRNPNAAEGAKNIAMVGKGIVYDCGG LALKPAAHMTNMKTDMGGSAGVFCAFIAVVRSMKMQRTHFSHIANISVTLCLAENAIGPH SYRNDDVVVMKSGKSVEVMNTDAEGRIVLGDGVCYATGEQDFVPDVLIDMATLTGAQGVA TGSKHAGVYASDAEAEKDMINAGLRSGDLCYPVLYCPEYHEEVYKSPCADMRNTANSPSS AGSSCGGYFVEQHLSERFRGPFVHVDMAYPSSNTAGATGYGVTLV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_110630.1 | 237 S | NTANSPSSA | 0.994 | unsp | LdBPK_110630.1 | 237 S | NTANSPSSA | 0.994 | unsp | LdBPK_110630.1 | 237 S | NTANSPSSA | 0.994 | unsp | LdBPK_110630.1 | 92 S | AVVRSMKMQ | 0.994 | unsp | LdBPK_110630.1 | 121 S | IGPHSYRND | 0.996 | unsp |