

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
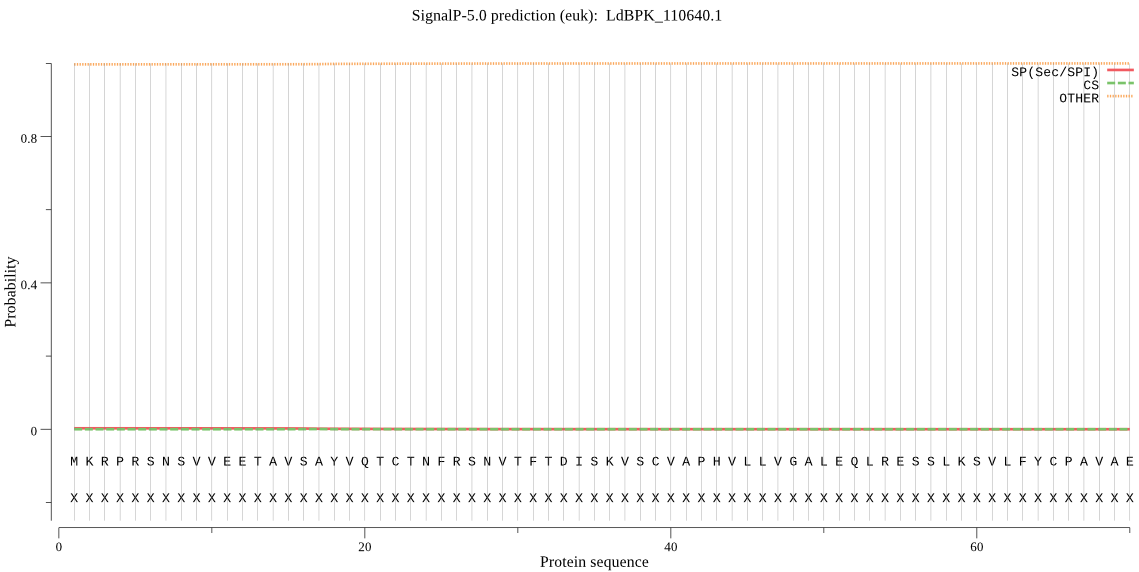
| LdBPK_110640.1 | OTHER | 0.998977 | 0.000317 | 0.000706 |
MKRPRSNSVVEETAVSAYVQTCTNFRSNVTFTDISKVSCVAPHVLLVGALEQLRESSLKS VLFYCPAVAEALQRVKAGATVKTLAEVPGRKGYTEVTVTALPATTSRTNCPYRADSMSEA VAAACGSVEEGEVLDVYVRAPAGSETAVANAVARAAPHSYTAKARQATKAYMKQAVTLNV VMSSRAAFTQELVRGKSVCVAELEAICTSVQLCQRLVDTPPCMLDTVVYAEIAAAYAAEL GVDMTVLKGEELREKGYGGIYAVGKCAQYPPHLVTLRYRNPNAAEGAKNIAMVGKGIVYD CGGLALKPAAHMTNMKTDMGGSAGVFCAFIAVVRSMKM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_110640.1 | 8 S | PRSNSVVEE | 0.997 | unsp | LdBPK_110640.1 | 116 S | YRADSMSEA | 0.998 | unsp |