

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
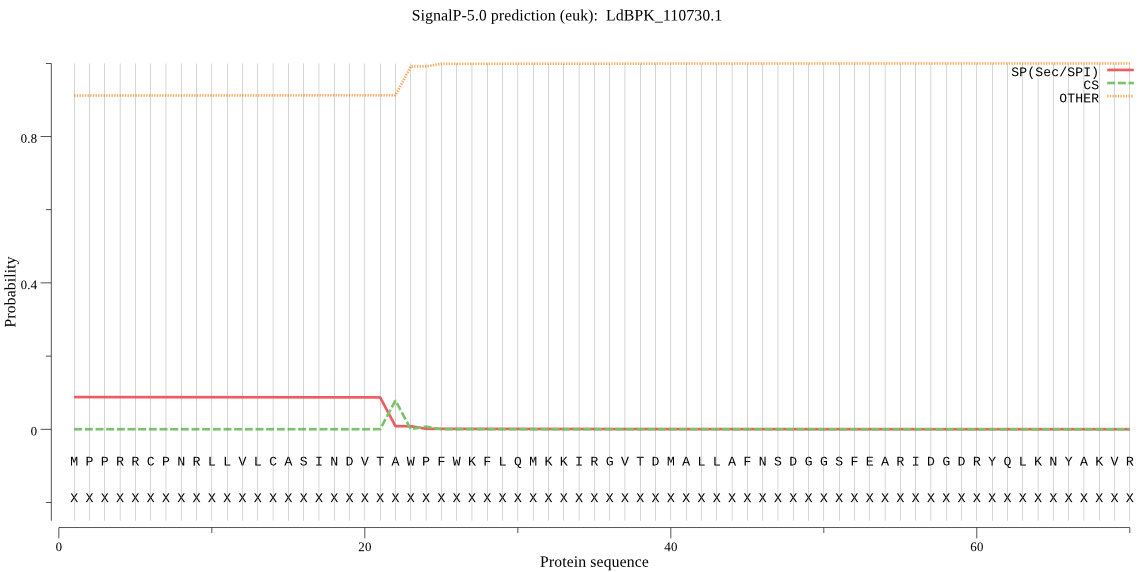
| LdBPK_110730.1 | SP | 0.447474 | 0.546509 | 0.006017 | CS pos: 22-23. VTA-WP. Pr: 0.9666 |
MPPRRCPNRLLVLCASINDVTAWPFWKFLQMKKIRGVTDMALLAFNSDGGSFEARIDGDR YQLKNYAKVRGYQHDMFESFVHRWHDPGRSYFVYGGHGMGDYVELEQNRVSLQAHELADV FGTRVFEAVLFDACFMANLDCAYHLRHNTRYIGACEGYMWEPDTALDYHVFNTHNASAMS RFKDPLHILRVIQADYCSKAPRGDFTIIDTTHIAALRQYVQEHVMQRVYDRATFYSLPQR ERLQQIAEASIQASISQFGHPGGDTNVMNGVGSGTGRPRTAPSSPEVLALSAAGRPTRRQ RMLQAIQFEHSLYPSEVDDKQLLDLKSYLTDMLREEQQLKAWEAAALGPQRRIAAGRRRL RSDGLQHGGSSGIAAPSSASSSFAVWKAPPSRALFVDRHGRLPAASSTGYSPSINGGPAA ANDKQKEATALSPPALAATLTMATAPPPSLSTSYKGSAQEGLDLFHQVVVSHIPPKAASI YATQLGGLSLTVHEYSAMSRPAEPWSVGSKRLLKRRAKQFLKNGELSEVVMESPKASPSV TSAALAATVNKATATAAAVKGASAGKPQHATSAAALPLSPRTRMLFPDQRLSFTSSSNGT NSDSSLPLSLSAPLSTSSTDACNSSMKTVILDPPQRAASCTSLPNSKQRTSAC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_110730.1 | 533 S | VVMESPKAS | 0.995 | unsp | LdBPK_110730.1 | 533 S | VVMESPKAS | 0.995 | unsp | LdBPK_110730.1 | 533 S | VVMESPKAS | 0.995 | unsp | LdBPK_110730.1 | 284 S | TAPSSPEVL | 0.996 | unsp | LdBPK_110730.1 | 453 S | SLSTSYKGS | 0.994 | unsp |