

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
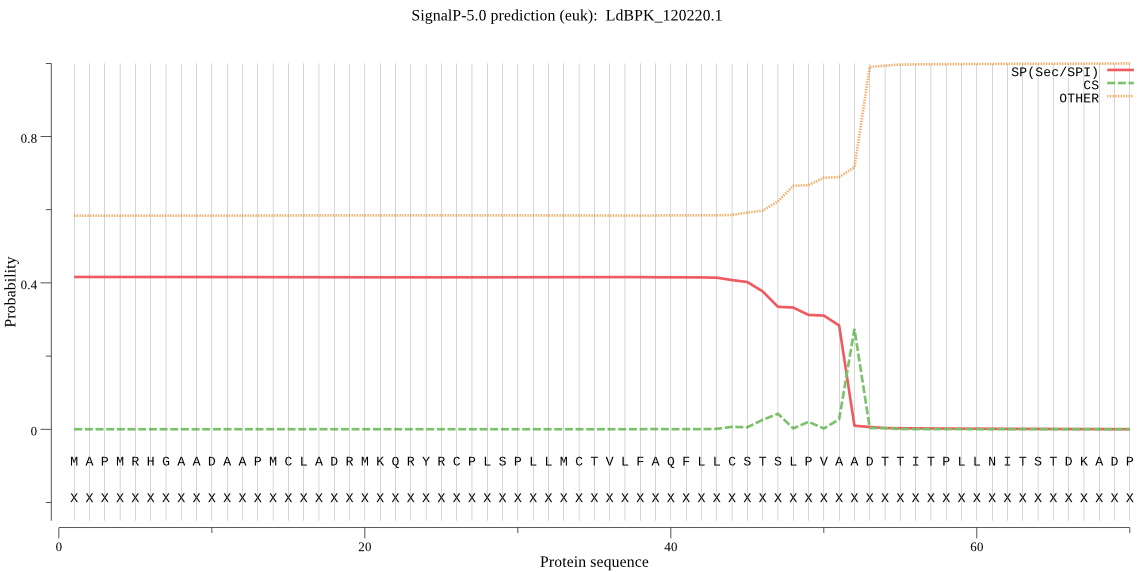
| LdBPK_120220.1 | SP | 0.482764 | 0.502685 | 0.014551 | CS pos: 52-53. VAA-DT. Pr: 0.6295 |
MAPMRHGAADAAPMCLADRMKQRYRCPLSPLLMCTVLFAQFLLCSTSLPVAADTTITPLL NITSTDKADPDEYYKILLSQDATILAGSTTVLSSDGVAYPNLNMVKGLRSVPQVPGAPMA QSAPKRTNTALKAFRADVLSEFLVAMALLYLDSTDALPFARTAPIPSTYLPASYTGGASF VNPSFPTVPITLNMLLQHVSSLTETGLDLRAETAPSGAVSTLSAFVSSLFSTTTTTVFSA SQPGLSTSYTYSHANVAIVAYVVEQVLATSTNYSALSGIGEFVFTVVMPPLQLSNTFLLN RNGHVIEAAYPLRSGSSHDTVNHVARQVLDSRNTGILDSAPREASYFSDYMLFTTSADVA KLAAEVLVPGGFYHMRLGASMLQNTVPIAVPTMRYTEARTTGLFLFSANKLCSTMYAAIS YSGQAPYCRYNSATVSESAPPFGLVATGGTDELAIVCVPVVSNTKTFCSIAELSFSGTGC SRCSPAAAGGRAVGLAMVSLAHLTSEPMPVAPAPTPQSTARTFNGWFVFLGVAVTLVVVV VASYITDYLIQPPPPAKIIAPVVAGQIGLRSGTALNGPVGGGGAVGSSEVPVDGESPEAF RENGRDHDAEDLYSGADDEEQTPYLARSGDRARGSSSLRRRCRRRRRRRHNGEPNPASDG SYSSGDDGWGDDRDSVDRVSTGSRRPSPKGALRFDAYI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_120220.1 | 622 T | DEEQTPYLA | 0.991 | unsp | LdBPK_120220.1 | 622 T | DEEQTPYLA | 0.991 | unsp | LdBPK_120220.1 | 622 T | DEEQTPYLA | 0.991 | unsp | LdBPK_120220.1 | 663 S | DGSYSSGDD | 0.992 | unsp | LdBPK_120220.1 | 675 S | DDRDSVDRV | 0.993 | unsp | LdBPK_120220.1 | 680 S | VDRVSTGSR | 0.994 | unsp | LdBPK_120220.1 | 687 S | SRRPSPKGA | 0.998 | unsp | LdBPK_120220.1 | 316 S | LRSGSSHDT | 0.997 | unsp | LdBPK_120220.1 | 596 S | VDGESPEAF | 0.995 | unsp |