

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
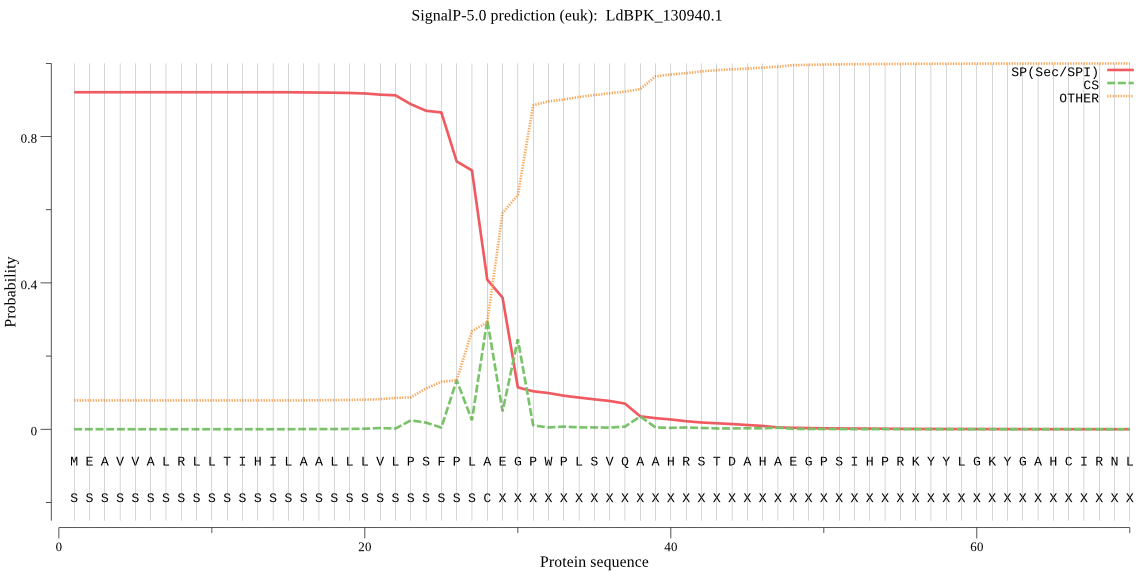
| LdBPK_130940.1 | SP | 0.035481 | 0.963290 | 0.001228 | CS pos: 28-29. PLA-EG. Pr: 0.4212 |
MEAVVALRLLTIHILAALLLVLPSFPLAEGPWPLSVQAAHRSTDAHAEGPSIHPRKYYLG KYGAHCIRNLTEFVASEDAAERLACFSGRGVRVALLETGLCAGIAERSRNSVTCTSVVPG VACEDAGCAHGTRSVSVLAGQLSVSAPLPREPLEETAWRNRETHLALADQYIGLAPDSTV RVLRIFDRQGRTNRRHLARALDLLLREAEDGEAGRLHSAWNTSSAHIRRRDETVDVISLS YGSKDYHSSPQVQDRLYRLMHEHGVIVVAAAGNDGARFGSVRSPADMPGVLAVGALRMEW HGRSAQTTQASRTLGDLASSLAAGGGHKSVAHFSGRGPTTWELPFGAGRAKPDLVALGQH VWAVQGVSAAAFSAAAPRARSASPALQLRSASGTSIAAPIVAGVVALCLEAAWSSTSAVA LAQNMTGGGSFLDVRQNRLARVSDSLRVREVILRTAVPLGEAAASLPPWPHSAPTTPLHS DSSSTASEAHPRRVLAGVPLLKLYAGYLRLSRVSILSQGAGEVQPLRALHAIVAKAAASS AIDAPEALRNCASFAIPTCVRIGYRIPRCSNSSSSSSSSSGDMSTPQSCQPPHDPDGKAH GDSPLPGRSGMSLDAPPAAYWWPFSDQAVYPGATPVLLNISLHLCPPSSPAAADQGTRDA AAAASGKGARRGSTTHIHVTHVITKVSGHVRAREGHPSRNSSHQCDACPLLFEGAKSRHR AGSTSPTRLLNLTARSASPRASCSDGEVAGEGATVMGPRHTQTAVCSSVKRWSTQQRWIH HLLRVATELTVVASRSQTRANVVEQLSPPPTSFSLSVAVSSPASAHTHLCYDVARETVAV WRKKGKSSPASEKDAAAGRRSADGDDGHDKQHSRDGAQVPADHDRCVPLFHLFRALSVEG ALHIFTGSSSSQPALTVPFAVRVVEPPPRVQRVLIDTSLDCFNPTTATSNLFIAGDDPHE SDAGGAGAALRRGRQRQRHERAYAEASGGDHPFTNLALLYLYLRHTLGMAVETFPLLHMS TIATSIAASHSPPLWPPAERTSNVSRTAQRAPAGTLAHIGTLIIVDPERPLTRGMRRLLT RAVLGGGADRSADGLNVLLVTDWYSADLAAQLHWARDESLYDAERFSHINGTDATDEVAG ADGDRDAVRDLRALRQSENGSTRGLAGSSHVPSWNRWLSEVTVASGSAANSSDCDGAAPL GSEGLLDGTSELPFELSESIVIDGAVLVDAAAPRRRSEPEGNAFASHGGTVTSAKVVALR SLGQLCAAGVLRWRLPAQAAVQRQAGRATAVGASKGVREPNVYPSGNGTCSTGDASNTSA MPSTQEGEAVICNVMPSWAQQQRRLVKRTVVSTRDGSAPAAVVEDGWEDVPVANGDGRLV RIGGLEAAEVSTMAPAATGASDATQSLTHGVLGFLTLPPSSTLVSTVSANRFRPGRIAIF TDSDCLSASDHHAQAALDELEALLYPPSASPSLPVTTCATWDCFSRTPDGQRLLQAESAQ SSVCVEVVKELLLWLHTGNLHRWRDSAQLQCEARTWARARHRSTADTTPGAGSVLEPKAG EDSVEGDRETANFDELLGTAPERAHVGEVVWRLWASLVESTERDAASFLVDGPKEGTRFQ GDYAAHQESAAANVMRALMVKYHGDWRAHLLDHNVSPPSTGAAGDSYSQSLRPARGRRSG IATVQQPEQHISLIESLLPLCGMLNALRWLQHPMVLGQLMVSAAVVMSVWLCRAGGGRWR GQRA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_130940.1 | 540 S | AAASSAIDA | 0.994 | unsp | LdBPK_130940.1 | 540 S | AAASSAIDA | 0.994 | unsp | LdBPK_130940.1 | 540 S | AAASSAIDA | 0.994 | unsp | LdBPK_130940.1 | 575 S | NSSSSSSSS | 0.991 | unsp | LdBPK_130940.1 | 577 S | SSSSSSSSG | 0.995 | unsp | LdBPK_130940.1 | 579 S | SSSSSSGDM | 0.997 | unsp | LdBPK_130940.1 | 673 S | ARRGSTTHI | 0.991 | unsp | LdBPK_130940.1 | 723 S | HRAGSTSPT | 0.997 | unsp | LdBPK_130940.1 | 725 S | AGSTSPTRL | 0.992 | unsp | LdBPK_130940.1 | 738 S | ARSASPRAS | 0.998 | unsp | LdBPK_130940.1 | 742 S | SPRASCSDG | 0.998 | unsp | LdBPK_130940.1 | 848 S | KGKSSPASE | 0.991 | unsp | LdBPK_130940.1 | 851 S | SSPASEKDA | 0.998 | unsp | LdBPK_130940.1 | 987 S | YAEASGGDH | 0.992 | unsp | LdBPK_130940.1 | 1119 S | ARDESLYDA | 0.996 | unsp | LdBPK_130940.1 | 1127 S | AERFSHING | 0.993 | unsp | LdBPK_130940.1 | 1237 S | PRRRSEPEG | 0.997 | unsp | LdBPK_130940.1 | 1323 S | SAMPSTQEG | 0.991 | unsp | LdBPK_130940.1 | 1352 S | RTVVSTRDG | 0.997 | unsp | LdBPK_130940.1 | 1543 S | ARHRSTADT | 0.997 | unsp | LdBPK_130940.1 | 1563 S | AGEDSVEGD | 0.993 | unsp | LdBPK_130940.1 | 1679 S | RGRRSGIAT | 0.995 | unsp | LdBPK_130940.1 | 111 S | RSRNSVTCT | 0.996 | unsp | LdBPK_130940.1 | 283 S | GSVRSPADM | 0.992 | unsp |