

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_161460.1 | OTHER | 0.994951 | 0.000761 | 0.004289 |
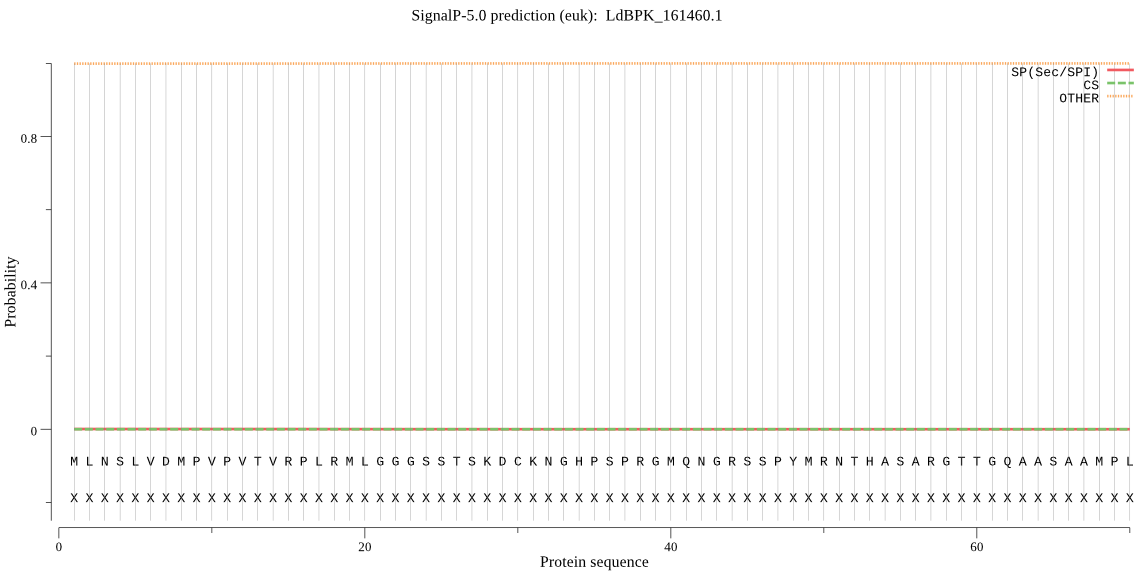
MLNSLVDMPVPVTVRPLRMLGGGSSTSKDCKNGHPSPRGMQNGRSSPYMRNTHASARGTT GQAASAAMPLVADPLTNMKSTVIMSSPTPPPPRPTITIFADLEDDVGAGGLSFGKASRAT VSSVAAGGSCRRVEHDPLTFTQTLLSPHGYSSLCQHAKAEVAAESAFSSSSERAGIEAMR RKSGSLLPAGGDRSPERICSGSFRVFSPADSAELAAQTDSSLPSGLPNDISWFHRRTTTY PGSEDAEPHPLRISATPVVPIVPHVAEPSSRKRVAEDMQRLYGGAGAAAGSIAVCSAKEA SQASTSVATRDVSWSSGCTGNTTDASQSPRLAAKKDKKLSLLTPFTSSASKKPTKVEDKT PPSTAADRRQAHEQIIRVGVQRLYQRLNELRLVVYRVRNDGNCQFRAISHQLFGNEDYHD IIRSQIVSYMRAARARSFDYYFESPAHADAYYHNLAKPGSWGDELSLRAASDCLYVNIHV LSSEERNCYITYRPSSDYAASAPSFLVDVWKLRERRRAERRLLRAYGPQQQQQQQGHSSS NAFGDSRQASLYAGPQGIGSDDEGEMDANAIQLALHRRLQQSEIRCSLPLSATGSYNSGG GGGGFSLGAATMPLLQPQSTTKKPSQLLTPAAVPVPRLSSGNEGASDDGAANQVAKFRAV GAEVQSLDKATQAVNIFTQRKESANAYNTNDVAVCFPRKHNEDATRPPFAGTASLELSRS SSPHPQAQHHGLTMSGSTTEEAPVSGGTRRADTMTSFQALSPTMRQGDEAGEVMLLASSM HLSRTTQSLQRSFSCMQRASSVTGPADHFNGLNARSSAHAPSAGGGESGASSQLLGSLVP RAATTSFRLGADDDGSLPSGSDAGSGANQGVCCFSFEPRTEPIDIFLSYLYPVHYNSLSV KQEQEPAAGTVPTAEATATPSTPPPQPSGNVSAR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_161460.1 | 207 S | FRVFSPADS | 0.997 | unsp | LdBPK_161460.1 | 207 S | FRVFSPADS | 0.997 | unsp | LdBPK_161460.1 | 207 S | FRVFSPADS | 0.997 | unsp | LdBPK_161460.1 | 238 T | HRRTTTYPG | 0.993 | unsp | LdBPK_161460.1 | 313 S | TRDVSWSSG | 0.996 | unsp | LdBPK_161460.1 | 350 S | TSSASKKPT | 0.997 | unsp | LdBPK_161460.1 | 460 S | AKPGSWGDE | 0.991 | unsp | LdBPK_161460.1 | 483 S | HVLSSEERN | 0.994 | unsp | LdBPK_161460.1 | 496 S | YRPSSDYAA | 0.997 | unsp | LdBPK_161460.1 | 550 S | SRQASLYAG | 0.993 | unsp | LdBPK_161460.1 | 722 S | SRSSSPHPQ | 0.995 | unsp | LdBPK_161460.1 | 801 S | QRASSVTGP | 0.995 | unsp | LdBPK_161460.1 | 183 S | MRRKSGSLL | 0.995 | unsp | LdBPK_161460.1 | 194 S | GGDRSPERI | 0.995 | unsp |