

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_180360.1 | SP | 0.001387 | 0.998483 | 0.000130 | CS pos: 30-31. AYA-AV. Pr: 0.4884 |
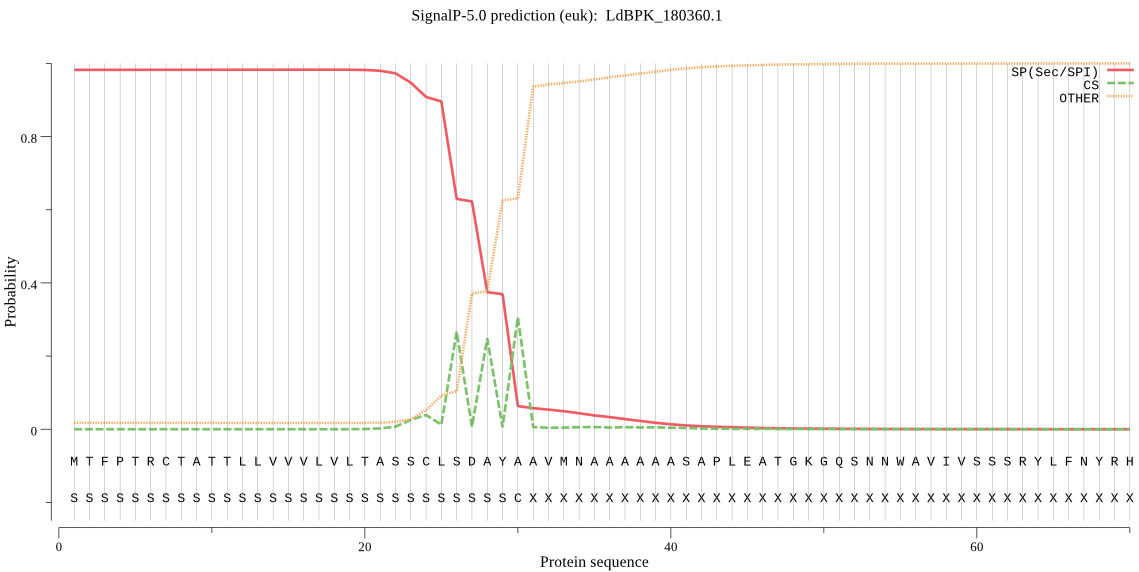
MTFPTRCTATTLLVVVLVLTASSCLSDAYAAVMNAAAAAASAPLEATGKGQSNNWAVIVS SSRYLFNYRHTANALTMYHLLRQHGIDDDHILLFLSDSFACDPRNVYPAEIFSQPPGERD ANGHASMNLYGCSAQVDYAGSDVDVRRFLSVLQGRYDENTPPTRRLLSDDKSNIIIYVAG HGAKSFFKFQDTEFLSSSDISETLMMMHQQRRYGRIVFLADTCHAIALCEHVEAPNVLCL AASDAESESYSFQYDEQLGTHMVSFWMNEMYSLLNGTSCSNPLTRRIGNDAVSVLHQSWY NFNYHPYRVEASRNRSKPAHRDAVNDPTALREWIVADFVCGRVPAAVPVDVRYDLE