

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_180450.1 | SP | 0.006001 | 0.993671 | 0.000328 | CS pos: 29-30. VYA-ST. Pr: 0.4709 |
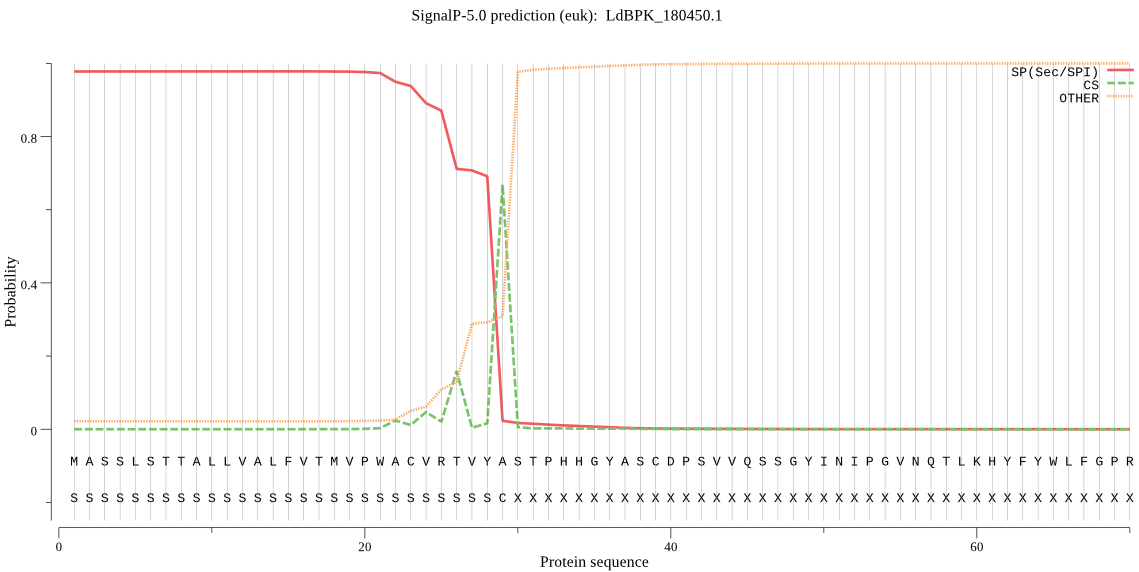
MASSLSTTALLVALFVTMVPWACVRTVYASTPHHGYASCDPSVVQSSGYINIPGVNQTLK HYFYWLFGPRKWPNDFREPPVIMWMTGGPGCSSSMALLTELGPCMMNETSGELYHNTYGW NDEAYLLFVDQPTGVGYSYGDKSNYAHNQSEVAEDMYNFLQLFARRFTSPSITGANDFYI IGESYGGHYVPAVSHRILMGNERSDGLHINLKGIAIGNGLTDPYTQLPFHAQTAYYWCKE KLGAPCITEKAYEEMLSLLPACLEKTKKCNEGPDDSDVSCSVATALWAEYVDHYYATGRN SYDIRKQCIGDLCYPMQNTIDFYHKPTVRASLGASAKAQWSTCNGEVSALFERDYMRNFN FTFPHMLDMGIRVLIYAGDMDFICNWLGNEAWVKALQWFGTDGFNTAPNVEFAVSGRWAG QERSYGGLSFVRIYDAGHMVPMDQPEVALFMVHRFLHDRRLA