

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_190120.1 | OTHER | 0.999554 | 0.000120 | 0.000325 |
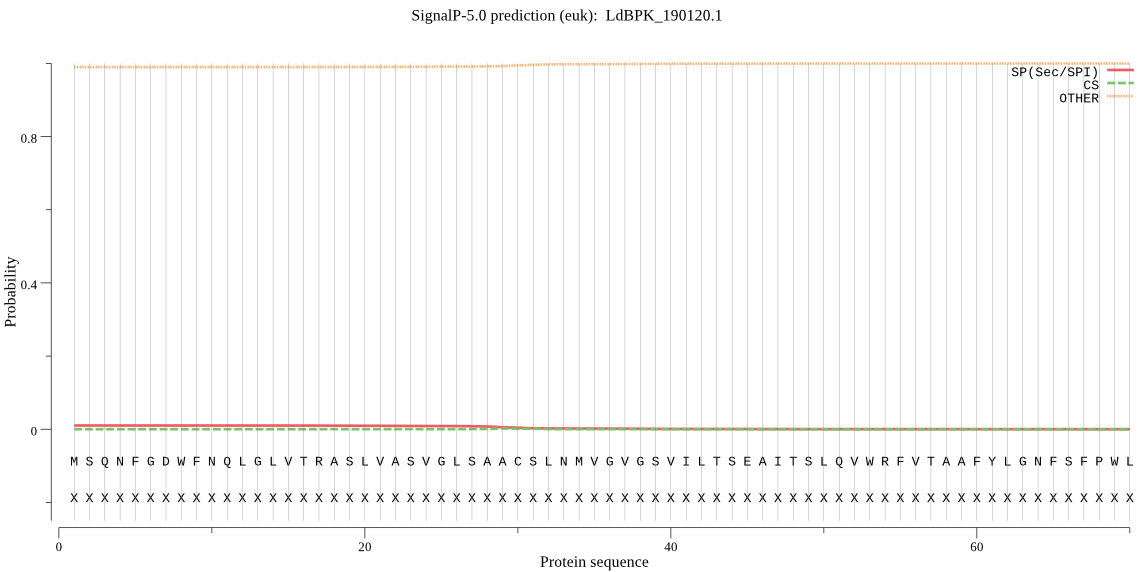
MSQNFGDWFNQLGLVTRASLVASVGLSAACSLNMVGVGSVILTSEAITSLQVWRFVTAAF YLGNFSFPWLMTVAMFVTYVKNNEESDFKGKTADMTYMFLLLVGALSSAGLFFNVYVTSF SFLMALCWIFCKRHPEQELTLFGFSFRSAVFPWVLMALHLVMGQGLLADILGIVAGHAYV FFKDVFPVSHNQRWLETPMWLRRQFTQPTHRVASFGPEMHPYDPRFQAAWRGEAQQRSSG LHNWGRGQTLGSS