

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_201720.1 | OTHER | 0.985084 | 0.011359 | 0.003557 |
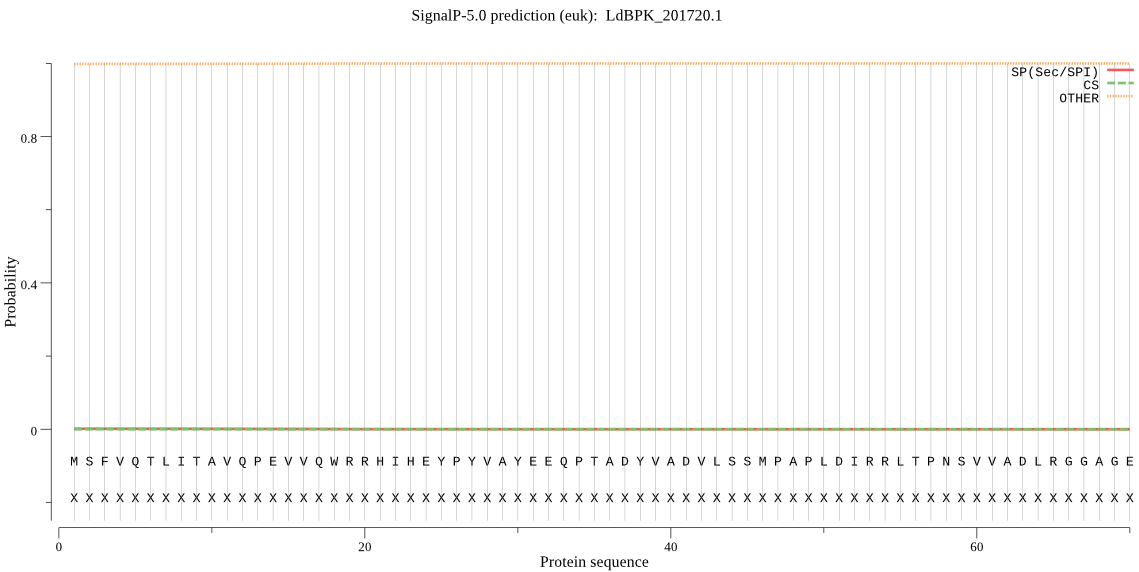
MSFVQTLITAVQPEVVQWRRHIHEYPYVAYEEQPTADYVADVLSSMPAPLDIRRLTPNSV VADLRGGAGEGPMYALRADMDALPLQEESGEPFSSKRPGVMHAC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_201720.1 | 56 T | IRRLTPNSV | 0.99 | unsp | LdBPK_201720.1 | 94 S | GEPFSSKRP | 0.992 | unsp |