

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
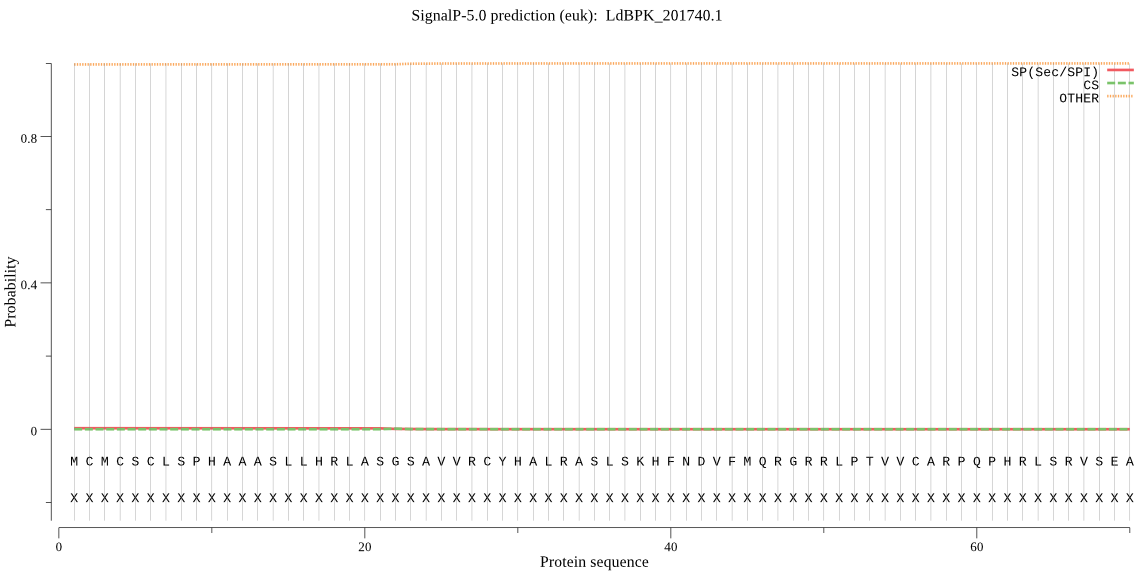
| LdBPK_201740.1 | mTP | 0.294544 | 0.005388 | 0.700069 | CS pos: 35-36. RAS-LS. Pr: 0.3324 |
MCMCSCLSPHAAASLLHRLASGSAVVRCYHALRASLSKHFNDVFMQRGRRLPTVVCARPQ PHRLSRVSEAPLLVLHFSALLPFLPRLLIHCTPDGALQRTPHSSCLHISPLWRLVWKLSL SLHENTWIVGEQIITHIPHSIVDKKTFYMQPFISFIDAVKDEVVQWRRHIHEYPYVAYEE QPTADYVADVLSSMPAPLDIRRLTPNSVVADLRGGAGEGPMYALRADMDALPLQEESGEP FSSKRPGVMHACGHDAHTAMLLGAVKVLCQMRD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_201740.1 | 242 S | GEPFSSKRP | 0.992 | unsp | LdBPK_201740.1 | 242 S | GEPFSSKRP | 0.992 | unsp | LdBPK_201740.1 | 242 S | GEPFSSKRP | 0.992 | unsp | LdBPK_201740.1 | 140 S | HIPHSIVDK | 0.996 | unsp | LdBPK_201740.1 | 204 T | IRRLTPNSV | 0.99 | unsp |