

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
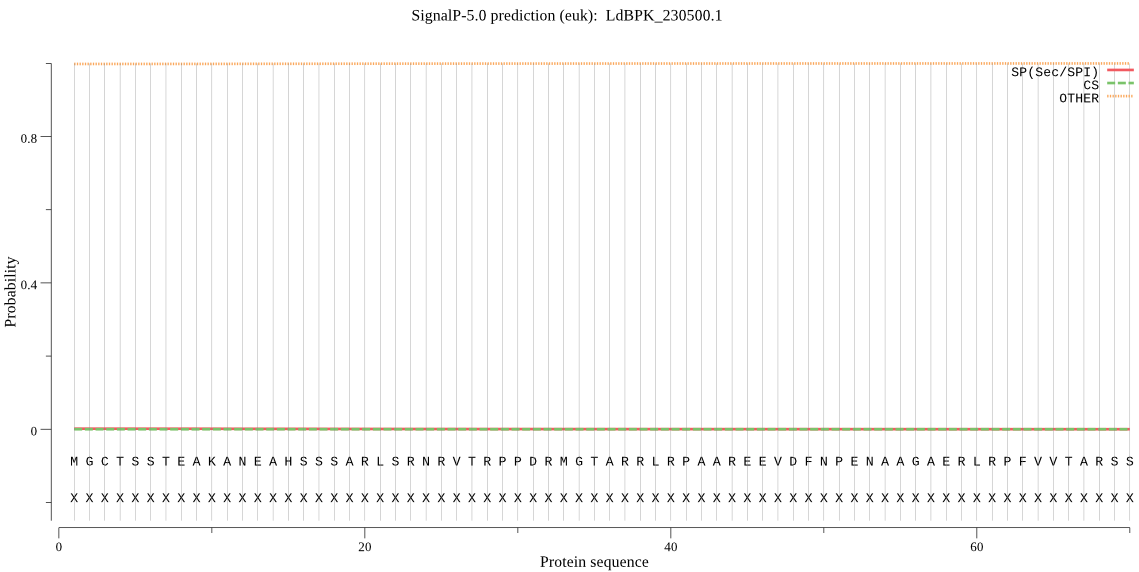
| LdBPK_230500.1 | OTHER | 0.999883 | 0.000027 | 0.000090 |
MGCTSSTEAKANEAHSSSARLSRNRVTRPPDRMGTARRLRPAAREEVDFNPENAAGAERL RPFVVTARSSSRVGAVTLHGVCGYTQNGVPAYSNGTSNTWTSTKNFREGIFMGYQWQCVE FARRWLWVTHRLLLPERSCAYCFSSCTYAYRLKKDPSPTSQRARAAMHGTAAETGAGSPS SFKEVNAATEVKLLGPYTNRDAWEKVPAKFVKQGSLVPPVADSLIVYPMSWGSPWGHIGV ITAVDLDRNCVYVADQNRYFHDWNGKSYSAVFRLDCDKGRFYIRDHESECMGWLTFPTAV EGHDVWCAGDQAPRKACKRQQEAGA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_230500.1 | 71 S | ARSSSRVGA | 0.994 | unsp | LdBPK_230500.1 | 157 S | KKDPSPTSQ | 0.994 | unsp |