

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
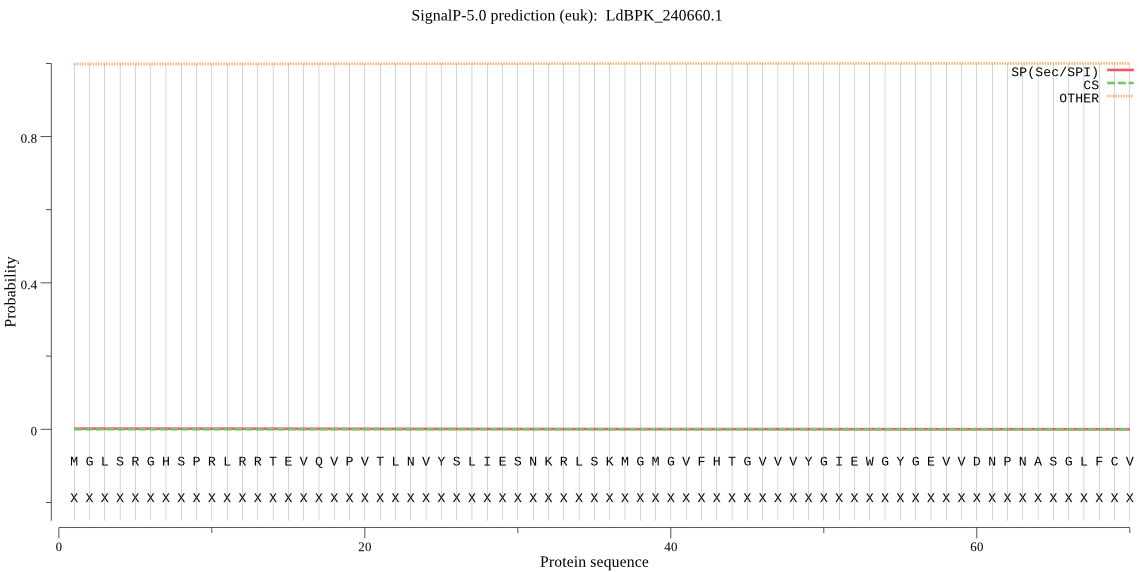
| LdBPK_240660.1 | OTHER | 0.996066 | 0.001045 | 0.002888 |
MGLSRGHSPRLRRTEVQVPVTLNVYSLIESNKRLSKMGMGVFHTGVVVYGIEWGYGEVVD NPNASGLFCVHPGQAAGTLYRTIRIGHTTRSPMQVDTILHRLENEWRSSDYHILHHNCNH FAQAFCDLLSTTEKLQVPSWCNRAARVGDRVIPRRLATKVQHMMDDEPPKAAAPPTPASN ISEVPTSVVPHEWYLHPSICQPLRYIDESKPLSVVTQDGHSASASVAAVDGGGSHYSAEC DIVPPPGYEMTDTGSVCPPITRRETYCSTDEKGSITHMMAIEEQPSVAPSSRRESACKST ASQRAAKTTSGHSPAPPSPGIAFSSGGEDRVVLPRMRASTSQSYSVAPTGSPQASEIEID VENVLQSETSEEPSPPVMSHLPMISSPNRGIVNPRKATPSGSTTSRQQGASFEQQHHVEN DSRDKGAEASSKKPLCMICSSSMESMSEELSDHGGSSAQPHSSFLATSNAMPVGCITTLL STSPNPSAEEATRRSSSMMAGERDVSTGVANMSSEVSNNTSGDFASVKHKHRTQATTTSS AIKEQKPKREPGNEFSDITGALVCDAPLSSATAEASPAGIKRKNSSAWKLLKNQCGLSSK TGSGNAVCKYLSTPSASLTSSSSFSLLQPEAHDDKEAVSVAADSGAVDPAADSALASRQE RGHMHVLLSASPSPPSPHSTPVALERKYSGHDAVSNEVQDANGTTPMPTTAALQNAPRVP HLGGDPPTSATELMPQSPEQRLSRSETSSAQGDVPSPVSPPDLDPATTASRPTDSPGPQL PPSFVTPERPNIITNSGAVTTGCSPAAAHQQCRRASSSAAMSDSPPPQRERCRTPIPFAG RRRGNDENVAPALRLDLDSDEAEPPVRRSNSRTAVEAGTPSTADKTAHAPMVPCSCGPSS LRREETATGDGRTGENASSDCSQAPLAVQPSLNTELADPTRLSPTPLESIPAPEANQPPT LPPCGTH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_240660.1 | 221 S | QDGHSASAS | 0.993 | unsp | LdBPK_240660.1 | 221 S | QDGHSASAS | 0.993 | unsp | LdBPK_240660.1 | 221 S | QDGHSASAS | 0.993 | unsp | LdBPK_240660.1 | 290 S | SVAPSSRRE | 0.991 | unsp | LdBPK_240660.1 | 291 S | VAPSSRRES | 0.997 | unsp | LdBPK_240660.1 | 295 S | SRRESACKS | 0.997 | unsp | LdBPK_240660.1 | 313 S | TSGHSPAPP | 0.992 | unsp | LdBPK_240660.1 | 355 S | SPQASEIEI | 0.994 | unsp | LdBPK_240660.1 | 370 S | QSETSEEPS | 0.991 | unsp | LdBPK_240660.1 | 445 S | SSMESMSEE | 0.994 | unsp | LdBPK_240660.1 | 447 S | MESMSEELS | 0.992 | unsp | LdBPK_240660.1 | 487 S | SPNPSAEEA | 0.997 | unsp | LdBPK_240660.1 | 496 S | TRRSSSMMA | 0.996 | unsp | LdBPK_240660.1 | 497 S | RRSSSMMAG | 0.997 | unsp | LdBPK_240660.1 | 676 S | PSPPSPHST | 0.996 | unsp | LdBPK_240660.1 | 689 S | ERKYSGHDA | 0.996 | unsp | LdBPK_240660.1 | 743 S | EQRLSRSET | 0.997 | unsp | LdBPK_240660.1 | 759 S | PSPVSPPDL | 0.996 | unsp | LdBPK_240660.1 | 869 S | PVRRSNSRT | 0.993 | unsp | LdBPK_240660.1 | 900 S | CGPSSLRRE | 0.991 | unsp | LdBPK_240660.1 | 943 S | PTRLSPTPL | 0.995 | unsp | LdBPK_240660.1 | 8 S | SRGHSPRLR | 0.994 | unsp | LdBPK_240660.1 | 35 S | NKRLSKMGM | 0.993 | unsp |