

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
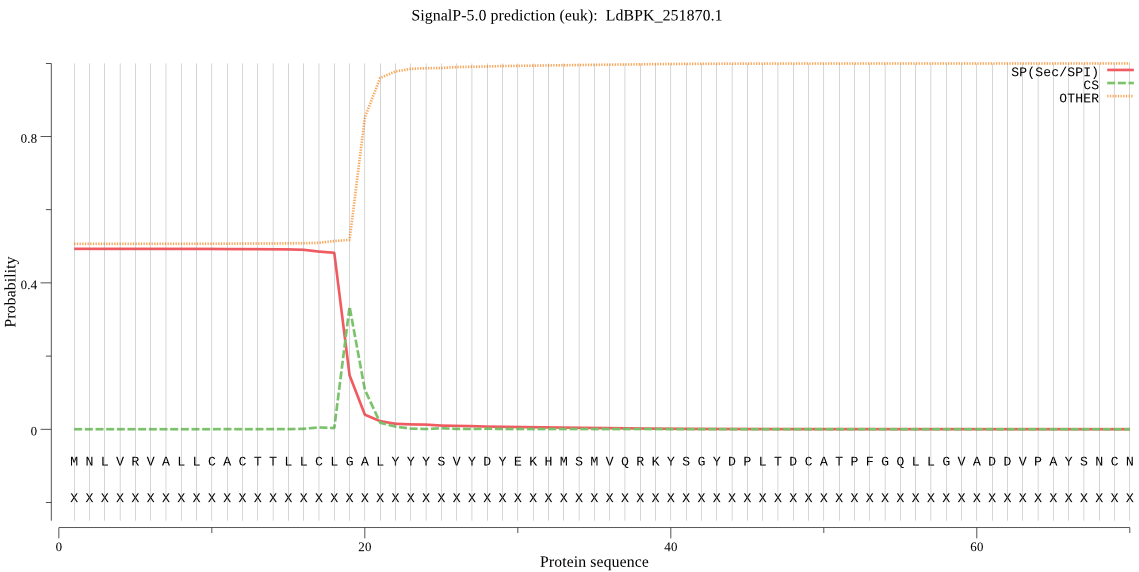
| LdBPK_251870.1 | SP | 0.033419 | 0.966528 | 0.000053 | CS pos: 19-20. CLG-AL. Pr: 0.6999 |
MNLVRVALLCACTTLLCLGALYYYSVYDYEKHMSMVQRKYSGYDPLTDCATPFGQLLGVA DDVPAYSNCNTKFSSTYINYVSLMDPMDNGRRGDPSETRIVMTAYRYTAFDYCMRWLVWN RGVMPRLVENTNQLWKTVDYFNPAKPEQGWSAEYITNYEEVTGMEERKFNAPRRADAIVY RMDKNTIPAGHIAVVVKVEDDVEAAGGPEKLKELKKMRLHPRRVYVAEQNWKNQPWGGHN YSRVLQFKWRAVAEKAYEGCYVDPDGLGIIGVVRVGKAMPLRAAPDPYQEALDMDNDGDL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_251870.1 | 41 S | QRKYSGYDP | 0.997 | unsp |