

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_262720.1 | OTHER | 0.610299 | 0.284822 | 0.104879 |
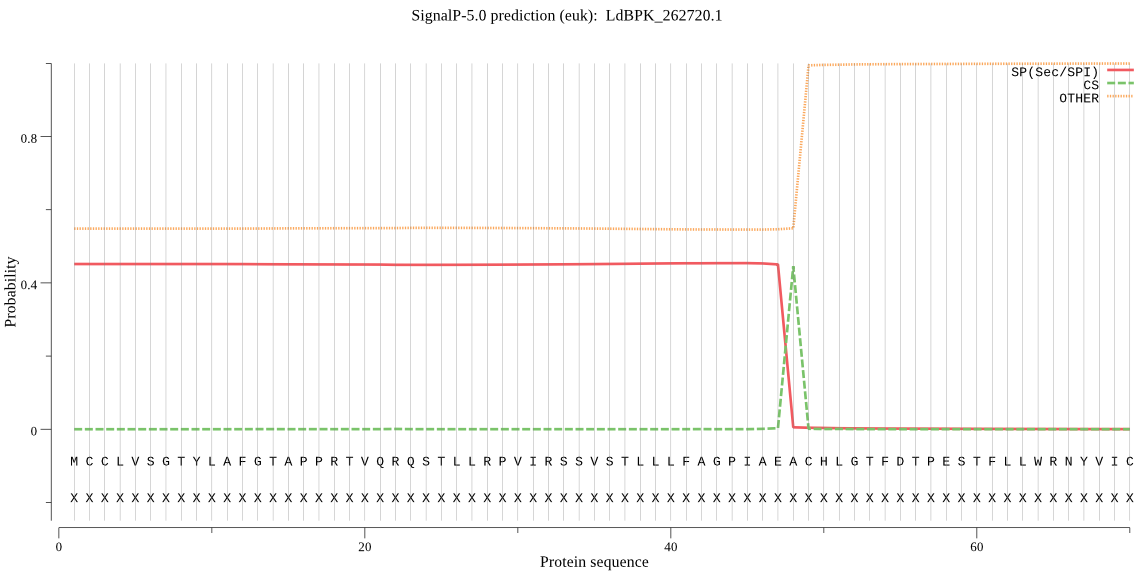
MCCLVSGTYLAFGTAPPRTVQRQSTLLRPVIRSSVSTLLLFAGPIAEACHLGTFDTPEST FLLWRNYVICPIGEELFYRGVLFSLLHRRSSAARIFVSAFLFSLSHIHHLVSMACDAYRN CEDKGVAGNDRDEKERACWRSAGHAMCGIFVFTALFGLLSGYYYEHVCERSIIAIAAAHA LCNAIGPPEFTVLRSRHFTAREKVASAAIYVAGIVGWAWTLLRY
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_262720.1 | 91 S | HRRSSAARI | 0.992 | unsp |