

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
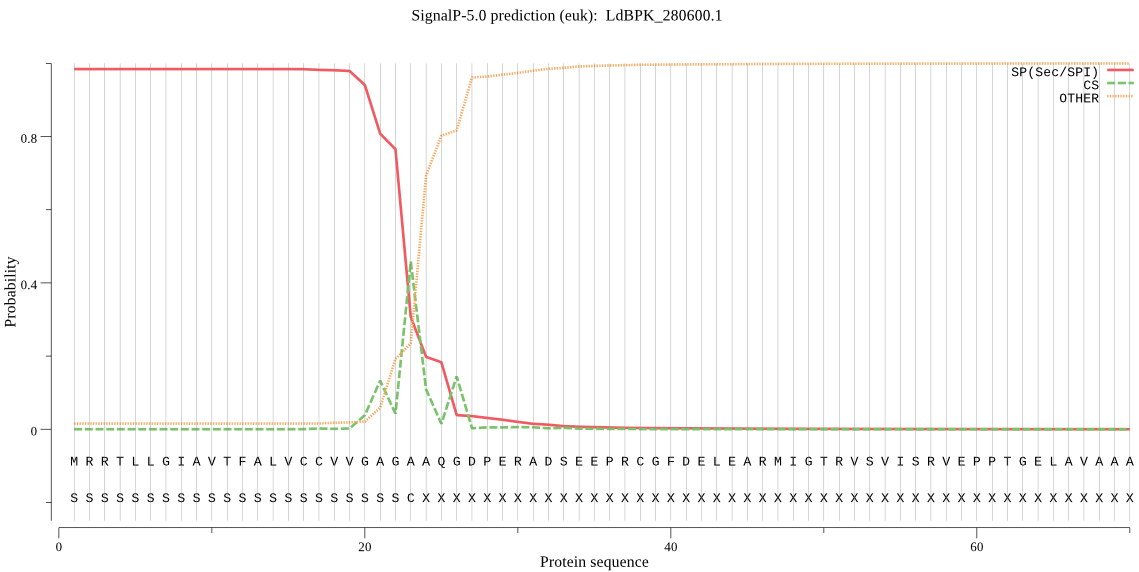
| LdBPK_280600.1 | SP | 0.001802 | 0.998180 | 0.000017 | CS pos: 23-24. AGA-AQ. Pr: 0.5099 |
MRRTLLGIAVTFALVCCVVGAGAAQGDPERADSEEPRCGFDELEARMIGTRVSVISRVEP PTGELAVAAAATGAWQPIRIAVFTEDISNSSQHCTASGQSRPTFRGGRVTCSAADVLTRE KKRVLLELLIPSAVQLHQERLNVQRENGNIVVSPFIKKHSICGQFSIPEEHMKTGVPDAD FVLYMSAAPTSGSVIAWAVKCQSFDNGRSSVGVATISPKYITAEPKTVRVVAHEVLHALG FTRSVFKQQNMLVMASFRGKSPSPVIRSANVVAQAQLHYGCKTQASMELEDEGGKGTVSS HWKRRSAKDELMAGFSGVGVYSALTIAAMEDTGYYQGNYAKAEPMAYGHEVGCKLSSERC VIKSTSQIPAMFCDAPDAPWSCTSDRRGIGRCILTSYNSNLPTYFQYFGDPRLGGPDPLM DFCPFVRAADDTMCAAKTNALKGSVYGVMSRCVDTPAGFSIDDSAVQQHGICAEVQCGSS AYGVKINGASAFRDCPPGSTYNLSTLSPSFSKGHLVCPSYESVCAININASLYEEYSRLL TDHSVTGARTSVTAVVAVLLVVLFMG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_280600.1 | 210 S | NGRSSVGVA | 0.993 | unsp | LdBPK_280600.1 | 210 S | NGRSSVGVA | 0.993 | unsp | LdBPK_280600.1 | 210 S | NGRSSVGVA | 0.993 | unsp | LdBPK_280600.1 | 306 S | WKRRSAKDE | 0.997 | unsp | LdBPK_280600.1 | 33 S | ERADSEEPR | 0.996 | unsp | LdBPK_280600.1 | 56 S | VSVISRVEP | 0.992 | unsp |