

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
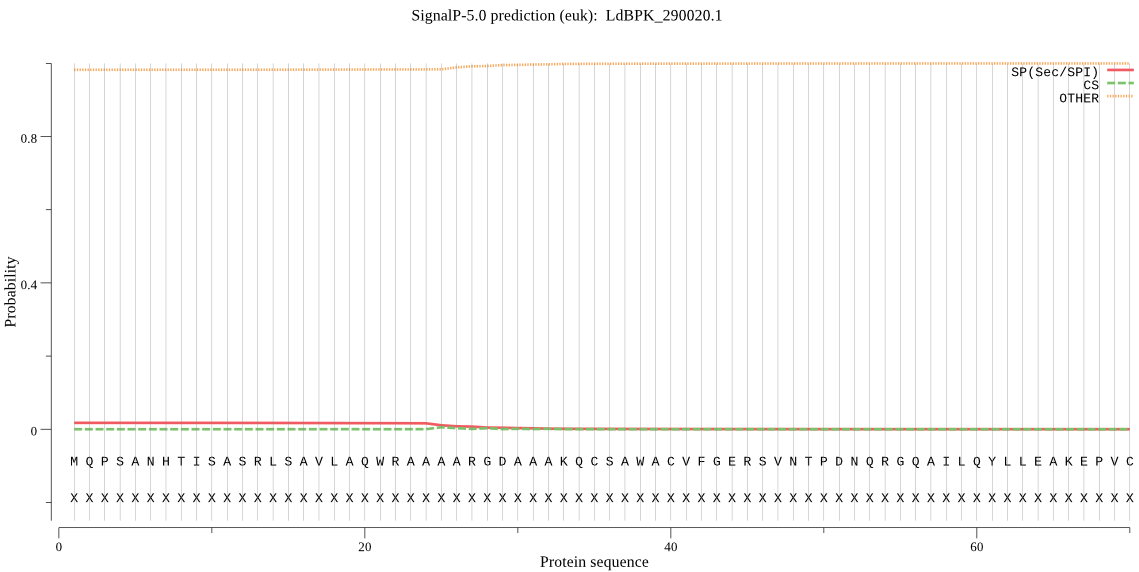
| LdBPK_290020.1 | OTHER | 0.762237 | 0.099827 | 0.137936 |
MQPSANHTISASRLSAVLAQWRAAAARGDAAAKQCSAWACVFGERSVNTPDNQRGQAILQ YLLEAKEPVCNTAVLITEDVTLILHGAGVRVAAPLTKLEGHTIEVVEGVEAAQARATALL AGKTVGAPVNEFAIQEGPFASAFTELVRGAVPSESLLSAAPVLGELLFVKDDAALGCVEK AAGLCCAVFRRYARDCIANEMSKRDPKTLYDVRQMLYAKLERPNTIQALESLAVDDFSLV TGLPPCLFHKGTYNSQLNVDEDTLKAACNVPIHGDVVVVRFGVKNTGYTAFFGRTLLVVS AAPSNAKEAYQFAYDVSAKLMELLVPGARLSDVYAGAMQYATNQNSDLAVLLVKNFGFST GLLVLEARGTISEKGTAIVADGMSFVIRVVLESVPSAGGKDTFDMELTDTVIIRGGVAEL KTKVARKLAEVLYEDLDETAAATEAAQQVRRDLSKITRQGQSDTVMISREAQREQELRQL LSELHAEFVAAGGKKGTQISTEEYRACDIGRLSLGELTPYKPEDRVPPPESKGGIFVQTD KKVVWLPVCGRAVPFHASTVNRVDVKTEGGKYTMTVTFHAMQEANVGYKLNPTKVFVKEL GYSSSRDVFTDSAIAIQGIQQRIKNEDAARKRALTSASNGRLTVTPNPLRLPTVKIRPPI TNANRQNKGCVGNLELHANGLRFSFLGGTPIDMLFENIKHVIFQPAVKSIYVIYHVTLKK PIEINRKNVLEVQFVAEVMESSELASAARRSFEEEVQAEERDEMRIRQTNKQFITFAHAV EERSKIKTQLPTNQFSFDGVHARSMTMFKGNREVLWAITDTPAFTQSVDEVEVVSFERII PGSATFDMSLILKDYNKPVITINSIPRDSLENIKDWCLSARLYYMETTVNPNWRATMKEI REDPDWDPWLRGDGWSVLNNVTNDEEEEEEDDDSDSDSTYYEDDDETSDSDDSSWLEDEE SDPSSGSDESDESSAASWDELERRAAAKDRQRDFSDDDDYHPRKRQRAGAATPALPVAPA TRPSKVPLNMAGRAGGGAVPPARRF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_290020.1 | 513 S | IGRLSLGEL | 0.995 | unsp | LdBPK_290020.1 | 513 S | IGRLSLGEL | 0.995 | unsp | LdBPK_290020.1 | 513 S | IGRLSLGEL | 0.995 | unsp | LdBPK_290020.1 | 751 S | AARRSFEEE | 0.998 | unsp | LdBPK_290020.1 | 869 S | IPRDSLENI | 0.997 | unsp | LdBPK_290020.1 | 934 S | EDDDSDSDS | 0.997 | unsp | LdBPK_290020.1 | 948 S | DDETSDSDD | 0.996 | unsp | LdBPK_290020.1 | 954 S | SDDSSWLED | 0.996 | unsp | LdBPK_290020.1 | 965 S | SDPSSGSDE | 0.997 | unsp | LdBPK_290020.1 | 967 S | PSSGSDESD | 0.992 | unsp | LdBPK_290020.1 | 970 S | GSDESDESS | 0.991 | unsp | LdBPK_290020.1 | 977 S | SSAASWDEL | 0.997 | unsp | LdBPK_290020.1 | 995 S | QRDFSDDDD | 0.998 | unsp | LdBPK_290020.1 | 202 S | ANEMSKRDP | 0.996 | unsp | LdBPK_290020.1 | 500 S | GTQISTEEY | 0.995 | unsp |